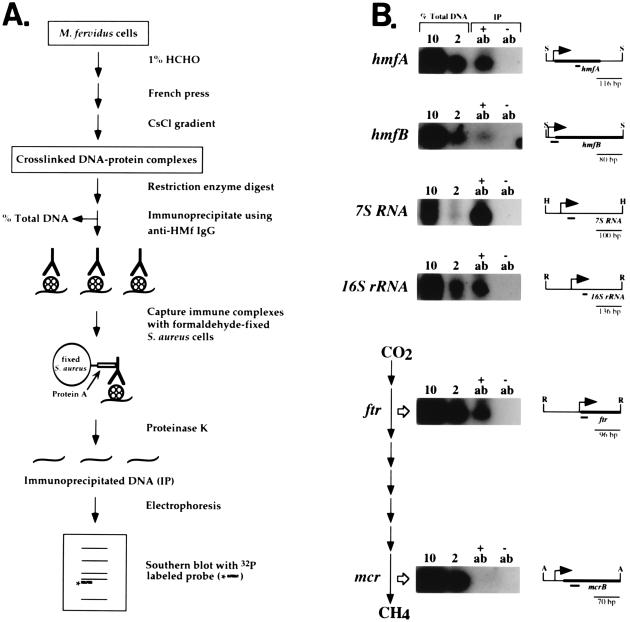
Figure 4.
(A) Outline of the procedure used to isolate and immunoprecipitate histone–DNA complexes cross-linked in vivo (20). (B) Southern blots generated with probes specific for the M. fervidus genes listed (27, 28). The probes (short heavy lines) were designed to hybridize, as indicated, to similar-sized restriction fragments, ApoI (A), HincII (H), RsaI (R), and SspI (S), that spanned the sites of transcription (bent arrow) and translation initiation (thick bar). As illustrated, the ftr and mcr genes encode the enzymes that catalyze the second and seventh steps in CO2 reduction to CH4 (27–29). At the time of formaldehyde fixation, the cells were actively synthesizing methane, and Northern blot analyses demonstrated that the ftr, mcr, hmfA, hmfB, 7S, and 16S transcripts were abundant. Aliquots (10% and 2%) of the total DNA present before the immunoprecipitation (see A) were electrophoresed in the tracks adjacent to the tracks that contained the DNA isolated from complexes immunoprecipitated (IP) by anti-HMf IgG (+ab) or by the control antiserum (−ab).