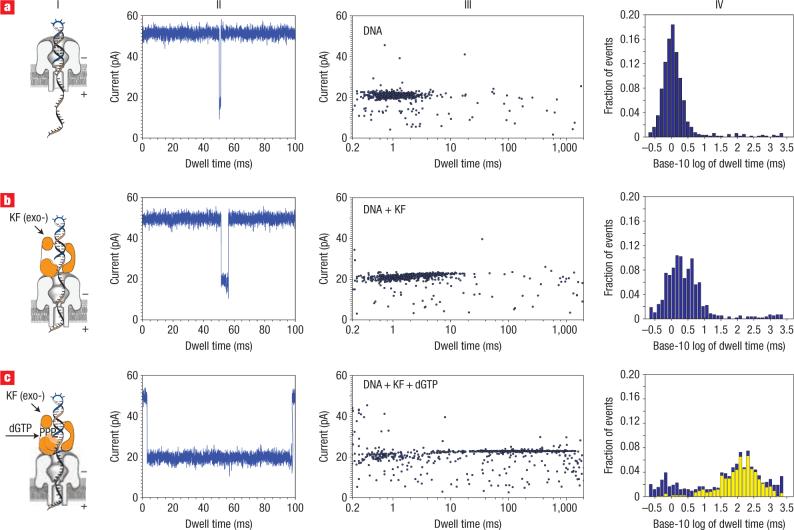
Figure 2. Distinguishing DNA, DNA/KF complexes or DNA/KF/dNTP complexes in the nanopore device.
a, Translocation, through the nanopore, of DNA alone (14-bp hairpin with a 36-nucleotide 5′ overhang and 2′–3′ dideoxycytidine terminus; template base at n = 0 is C). b,c, Translocation of the 14-bphp from complexes with KF (b) or from complexes with KF and dGTP (c). For each of rows a–c are presented a diagram of the nanopore with the associated complex (column I), a current trace (column II) and a dwell time event plot (column III). In column IV, probability histograms of the base-10 logarithm of dwell time data are shown in blue. Close examination of the event plot in c, column III, reveals that most long dwell time events are within 22 to 24 pA. A yellow subset histogram for the events within 22−24 pA is overlaid on probability histogram c, revealing that the chosen range is dominated by long dwell time events. The number of events, mean and IQR values for probability histograms a and b, and subset histogram c, are reported in Table 1.