Fig. 9. Molecular interactions formed by various human NAT2 residues changed as a result of NAT2 SNPs.
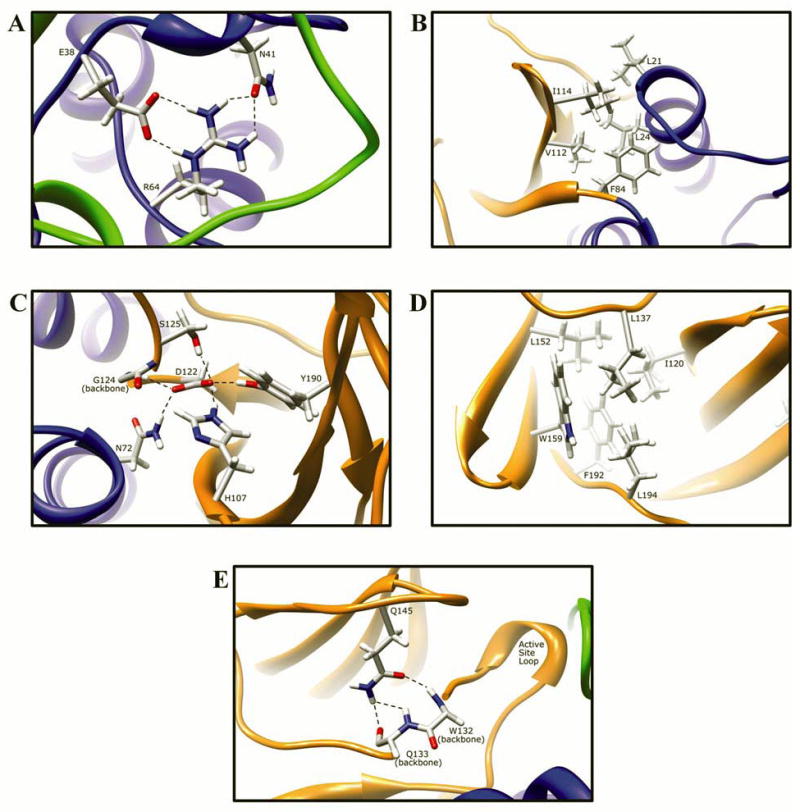
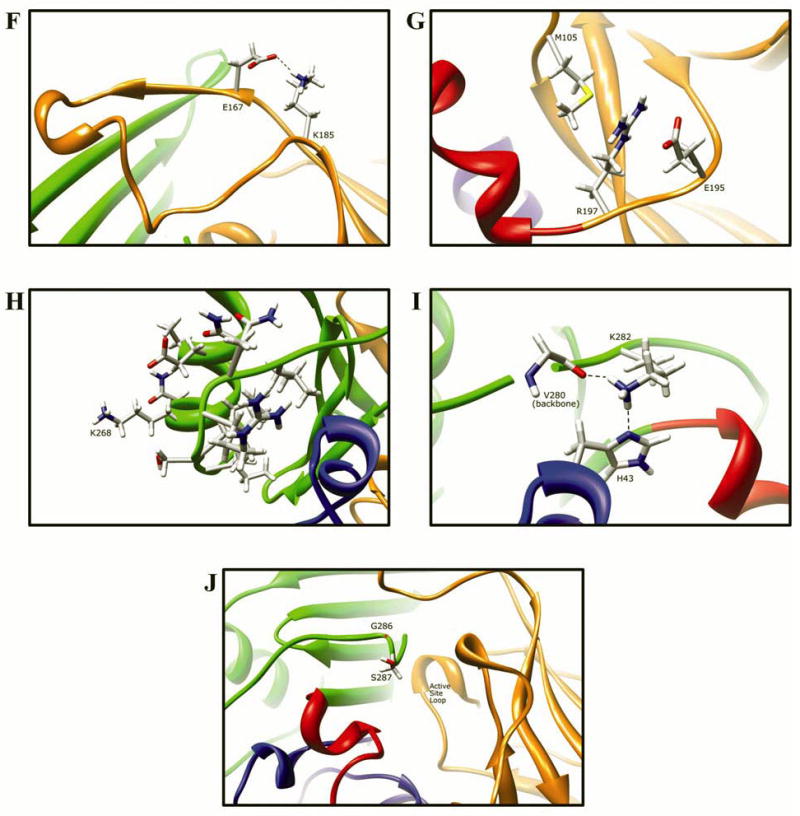
Protein domains are indicated by ribbon color as in Fig. 8, with backbone atoms shown instead of ribbon where necessary. (A) Residue R64 hydrogen bonds to E38 and N41 in domain I. (B) I114 is part of a hydrophobic core at an interface between the domain II beta barrel and a domain I helix. (C) Catalytic core residue D122 hydrogen bonds to N72, H107, S125, Y190, and the backbone of G124. (D) L137 is part of a hydrophobic core in the center of the domain II beta barrel. (E) Q145 hydrogen bonds to the backbone of residues Q133 and W132 in domain II. (F) Domain II loop residue E167 forms a relatively weak hydrogen bond to loop residue K185. (G) Positively charged R197 in domain I forms electrostatic interactions with negatively charged E195 and the M105 sulfur. (H) The side-chain of K268 is surface exposed in domain III, and does not interact with any other residue. (I) Domain III residue K282 hydrogen bonds to H43 in domain I and to the backbone of V280. (J) G286 is located adjacent to the active site pocket in the C-terminal region of domain III and has no interactions with other residues.
