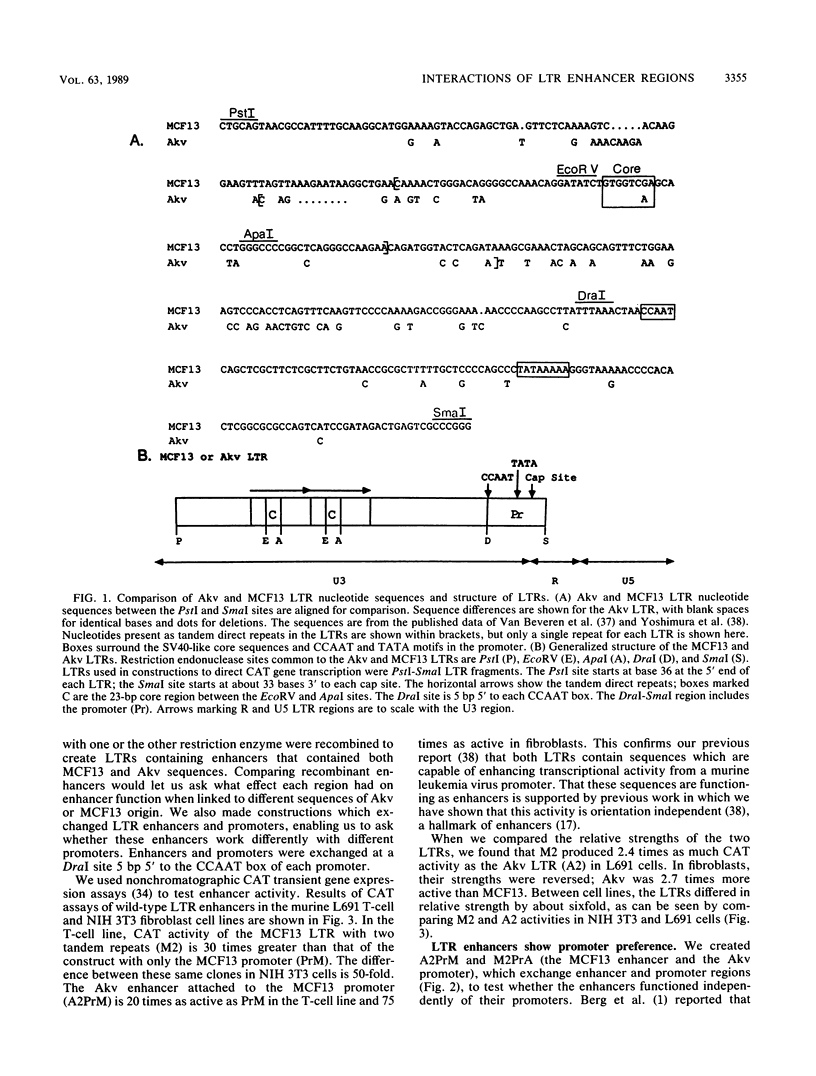
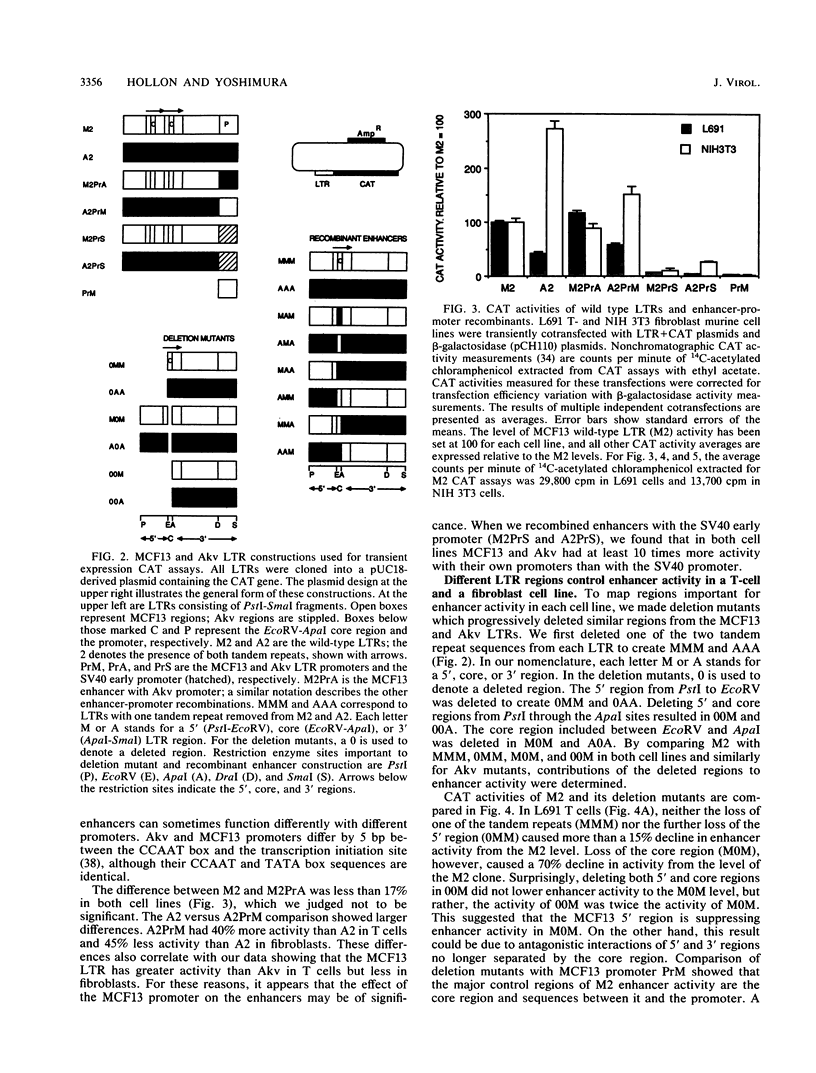
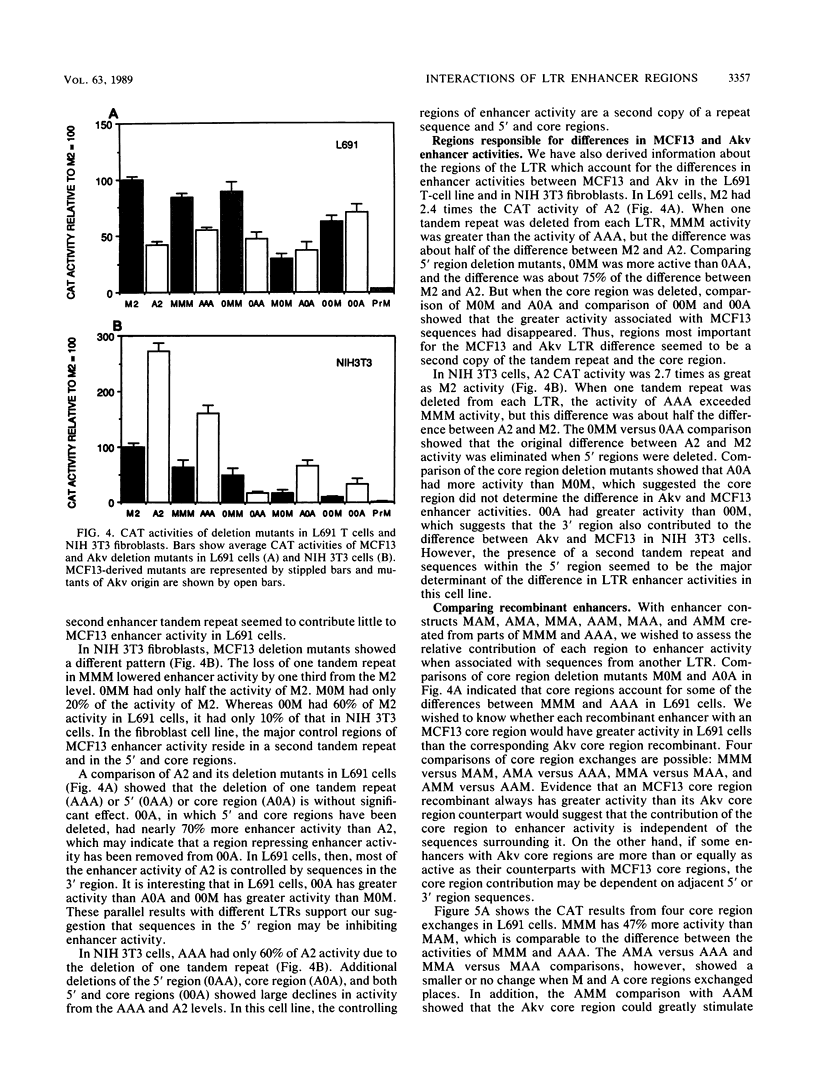
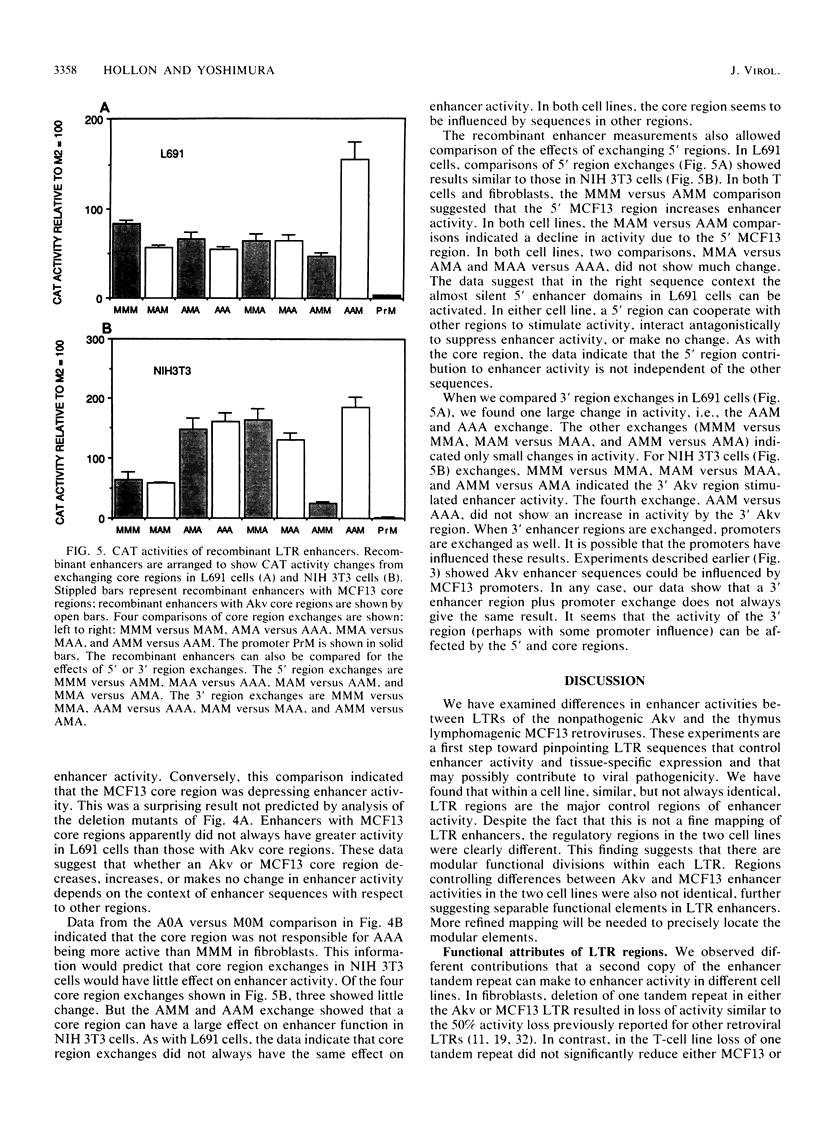
Abstract
We have used deletion and recombinant long terminal repeat (LTR) mutants to examine enhancer activity differences between LTRs of the nonpathogenic Akv and the thymus lymphomagenic MCF13 murine retroviruses. Deletion mutant analysis revealed that major control regions for MCF13 and Akv LTR enhancer activity were similar but not identical. For both LTRs, major control regions were distinctly different in a murine T-cell and a fibroblast cell line. Recombinant enhancer analysis showed that LTRs could be divided into three regions capable of altering the level of enhancer activity through cooperative or antagonistic interaction. The contribution of each region to enhancer activity was dependent on its context with respect to the other regions. LTR enhancer function in different cell types appears to be the result of the interaction of enhancer modular elements.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berg P. E., Popovic Z., Anderson W. F. Promoter dependence of enhancer activity. Mol Cell Biol. 1984 Aug;4(8):1664–1668. doi: 10.1128/mcb.4.8.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bösze Z., Thiesen H. J., Charnay P. A transcriptional enhancer with specificity for erythroid cells is located in the long terminal repeat of the Friend murine leukemia virus. EMBO J. 1986 Jul;5(7):1615–1623. doi: 10.1002/j.1460-2075.1986.tb04404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celander D., Haseltine W. A. Tissue-specific transcription preference as a determinant of cell tropism and leukaemogenic potential of murine retroviruses. Nature. 1984 Nov 8;312(5990):159–162. doi: 10.1038/312159a0. [DOI] [PubMed] [Google Scholar]
- Crabb D. W., Dixon J. E. A method for increasing the sensitivity of chloramphenicol acetyltransferase assays in extracts of transfected cultured cells. Anal Biochem. 1987 May 15;163(1):88–92. doi: 10.1016/0003-2697(87)90096-0. [DOI] [PubMed] [Google Scholar]
- DeFranco D., Yamamoto K. R. Two different factors act separately or together to specify functionally distinct activities at a single transcriptional enhancer. Mol Cell Biol. 1986 Apr;6(4):993–1001. doi: 10.1128/mcb.6.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L., Jolicoeur P. Mapping the viral sequences conferring leukemogenicity and disease specificity in Moloney and amphotropic murine leukemia viruses. J Virol. 1984 Nov;52(2):448–456. doi: 10.1128/jvi.52.2.448-456.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L., Jolicoeur P. The tandem direct repeats within the long terminal repeat of murine leukemia viruses are the primary determinant of their leukemogenic potential. J Virol. 1984 Dec;52(3):945–952. doi: 10.1128/jvi.52.3.945-952.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L., Rassart E., Jolicoeur P. Thymotropism of murine leukemia virus is conferred by its long terminal repeat. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4203–4207. doi: 10.1073/pnas.80.14.4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves B. J., Eisenberg S. P., Coen D. M., McKnight S. L. Alternate utilization of two regulatory domains within the Moloney murine sarcoma virus long terminal repeat. Mol Cell Biol. 1985 Aug;5(8):1959–1968. doi: 10.1128/mcb.5.8.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves B. J., Eisenman R. N., McKnight S. L. Delineation of transcriptional control signals within the Moloney murine sarcoma virus long terminal repeat. Mol Cell Biol. 1985 Aug;5(8):1948–1958. doi: 10.1128/mcb.5.8.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C. V., Jacob P. E., Ringold G. M., Lee F. Expression and regulation of Escherichia coli lacZ gene fusions in mammalian cells. J Mol Appl Genet. 1983;2(1):101–109. [PubMed] [Google Scholar]
- Holland C. A., Wozney J., Chatis P. A., Hopkins N., Hartley J. W. Construction of recombinants between molecular clones of murine retrovirus MCF 247 and Akv: determinant of an in vitro host range property that maps in the long terminal repeat. J Virol. 1985 Jan;53(1):152–157. doi: 10.1128/jvi.53.1.152-157.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimoto A., Adachi A., Sakai K., Matsuyama M. Long terminal repeat of Friend-MCF virus contains the sequence responsible for erythroid leukemia. Virology. 1985 Feb;141(1):30–42. doi: 10.1016/0042-6822(85)90180-1. [DOI] [PubMed] [Google Scholar]
- Ishimoto A., Takimoto M., Adachi A., Kakuyama M., Kato S., Kakimi K., Fukuoka K., Ogiu T., Matsuyama M. Sequences responsible for erythroid and lymphoid leukemia in the long terminal repeats of Friend-mink cell focus-forming and Moloney murine leukemia viruses. J Virol. 1987 Jun;61(6):1861–1866. doi: 10.1128/jvi.61.6.1861-1866.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones N. C., Rigby P. W., Ziff E. B. Trans-acting protein factors and the regulation of eukaryotic transcription: lessons from studies on DNA tumor viruses. Genes Dev. 1988 Mar;2(3):267–281. doi: 10.1101/gad.2.3.267. [DOI] [PubMed] [Google Scholar]
- Khoury G., Gruss P. Enhancer elements. Cell. 1983 Jun;33(2):313–314. doi: 10.1016/0092-8674(83)90410-5. [DOI] [PubMed] [Google Scholar]
- Koltunow A. M., Gregg K., Rogers G. E. Promoter efficiency depends upon intragenic sequences. Nucleic Acids Res. 1987 Oct 12;15(19):7795–7807. doi: 10.1093/nar/15.19.7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins L. A., Gruss P., Pozzatti R., Khoury G. Characterization of enhancer elements in the long terminal repeat of Moloney murine sarcoma virus. J Virol. 1984 Jan;49(1):183–189. doi: 10.1128/jvi.49.1.183-189.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins L. A., Khoury G., Gorman C., Howard B., Gruss P. Host-specific activation of transcription by tandem repeats from simian virus 40 and Moloney murine sarcoma virus. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6453–6457. doi: 10.1073/pnas.79.21.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenardo M., Pierce J. W., Baltimore D. Protein-binding sites in Ig gene enhancers determine transcriptional activity and inducibility. Science. 1987 Jun 19;236(4808):1573–1577. doi: 10.1126/science.3109035. [DOI] [PubMed] [Google Scholar]
- Lenz J., Celander D., Crowther R. L., Patarca R., Perkins D. W., Haseltine W. A. Determination of the leukaemogenicity of a murine retrovirus by sequences within the long terminal repeat. 1984 Mar 29-Apr 4Nature. 308(5958):467–470. doi: 10.1038/308467a0. [DOI] [PubMed] [Google Scholar]
- Li Y., Golemis E., Hartley J. W., Hopkins N. Disease specificity of nondefective Friend and Moloney murine leukemia viruses is controlled by a small number of nucleotides. J Virol. 1987 Mar;61(3):693–700. doi: 10.1128/jvi.61.3.693-700.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath M. S., Pillemer E., Kooistra D., Weissman I. L. The role of MuLV receptors on T-lymphoma cells in lymphoma cell proliferation. Contemp Top Immunobiol. 1980;11:157–184. doi: 10.1007/978-1-4684-3701-0_5. [DOI] [PubMed] [Google Scholar]
- Nagata K., Guggenheimer R. A., Enomoto T., Lichy J. H., Hurwitz J. Adenovirus DNA replication in vitro: identification of a host factor that stimulates synthesis of the preterminal protein-dCMP complex. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6438–6442. doi: 10.1073/pnas.79.21.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak T. J., Rothenberg E. V. Differential transient and long-term expression of DNA sequences introduced into T-lymphocyte lines. DNA. 1986 Dec;5(6):439–451. doi: 10.1089/dna.1.1986.5.439. [DOI] [PubMed] [Google Scholar]
- Ondek B., Gloss L., Herr W. The SV40 enhancer contains two distinct levels of organization. Nature. 1988 May 5;333(6168):40–45. doi: 10.1038/333040a0. [DOI] [PubMed] [Google Scholar]
- Potter H., Weir L., Leder P. Enhancer-dependent expression of human kappa immunoglobulin genes introduced into mouse pre-B lymphocytes by electroporation. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7161–7165. doi: 10.1073/pnas.81.22.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochford R., Campbell B. A., Villarreal L. P. A pancreas specificity results from the combination of polyomavirus and Moloney murine leukemia virus enhancer. Proc Natl Acad Sci U S A. 1987 Jan;84(2):449–453. doi: 10.1073/pnas.84.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirm S., Jiricny J., Schaffner W. The SV40 enhancer can be dissected into multiple segments, each with a different cell type specificity. Genes Dev. 1987 Mar;1(1):65–74. doi: 10.1101/gad.1.1.65. [DOI] [PubMed] [Google Scholar]
- Schulze F., Boehnlein E., Gruss P. Mutational analyses of the Moloney murine sarcoma virus enhancer. DNA. 1985 Jun;4(3):193–202. doi: 10.1089/dna.1985.4.193. [DOI] [PubMed] [Google Scholar]
- Sen R., Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986 Aug 29;46(5):705–716. doi: 10.1016/0092-8674(86)90346-6. [DOI] [PubMed] [Google Scholar]
- Sleigh M. J. A nonchromatographic assay for expression of the chloramphenicol acetyltransferase gene in eucaryotic cells. Anal Biochem. 1986 Jul;156(1):251–256. doi: 10.1016/0003-2697(86)90180-6. [DOI] [PubMed] [Google Scholar]
- Speck N. A., Baltimore D. Six distinct nuclear factors interact with the 75-base-pair repeat of the Moloney murine leukemia virus enhancer. Mol Cell Biol. 1987 Mar;7(3):1101–1110. doi: 10.1128/mcb.7.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiesen H. J., Bösze Z., Henry L., Charnay P. A DNA element responsible for the different tissue specificities of Friend and Moloney retroviral enhancers. J Virol. 1988 Feb;62(2):614–618. doi: 10.1128/jvi.62.2.614-618.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Beveren C., Rands E., Chattopadhyay S. K., Lowy D. R., Verma I. M. Long terminal repeat of murine retroviral DNAs: sequence analysis, host-proviral junctions, and preintegration site. J Virol. 1982 Feb;41(2):542–556. doi: 10.1128/jvi.41.2.542-556.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura F. K., Davison B., Chaffin K. Murine leukemia virus long terminal repeat sequences can enhance gene activity in a cell-type-specific manner. Mol Cell Biol. 1985 Oct;5(10):2832–2835. doi: 10.1128/mcb.5.10.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenke M., Grundström T., Matthes H., Wintzerith M., Schatz C., Wildeman A., Chambon P. Multiple sequence motifs are involved in SV40 enhancer function. EMBO J. 1986 Feb;5(2):387–397. doi: 10.1002/j.1460-2075.1986.tb04224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


