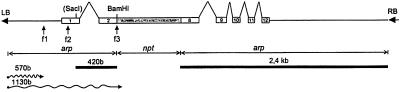
Figure 1.
A schematic drawing of the T-DNA from the pNPTARP binary vector. The left and right borders of the T-DNA are indicated as triangles. The arrowed lines below the T-DNA map indicate sequences derived from the arp gene or the nptII gene, which lacks the first eight amino acids. Boxed regions are exons. The arp gene exons are numbered and the nptII gene region is shaded. Appropriate frames are indicated as f1, f2, and f3. At the f3 site is an in-frame fusion of the arp second exon-coding frame with the coding region of nptII. At this position a BamHI site was generated. The beginning of the arp-coding frame with the initiating translation ATG codon is indicated as f2, and a 24-bp downstream frame shift (indicated in brackets) was introduced by deleting a SacI site. The frame shift results in an arp–nptII fusion, the frame of which begins at the f1 site, but lacks an ATG start codon. If the T-DNA integrates into the Arabidopsis genome by homologous recombination with the arp gene over 420-bp upstream and 2.4-kb downstream sequences from nptII (areas indicated by black bars below T-DNA), the nptII protein can be synthesized from the arp first ATG codon. In this case, the SacI site will be derived from a genomic arp gene and detection of the 420-bp SacI–BamHI fragment on a DNA gel blots can be used to identify homologous recombination events. Alternatively, before stable integration into the Arabidopsis genome, the T-DNA may undergo deletions, from 570 bp to reach the f1 site and up to 1,130 bp to reach the nptII-coding region, as indicated by waved lines below the map. In this case the expression of nptII may occur as a result of T-DNA integration into Arabidopsis-expressed genomic regions, given that an in vivo fusion protein will be synthesized in transformed cells as result of either direct exon-exon integrations or after splicing of the generated chimeric intron.