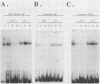
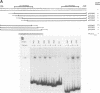
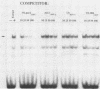
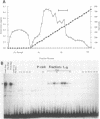
Abstract
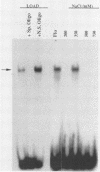
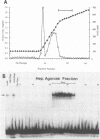
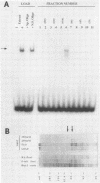
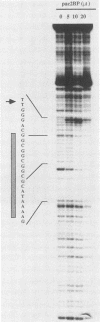
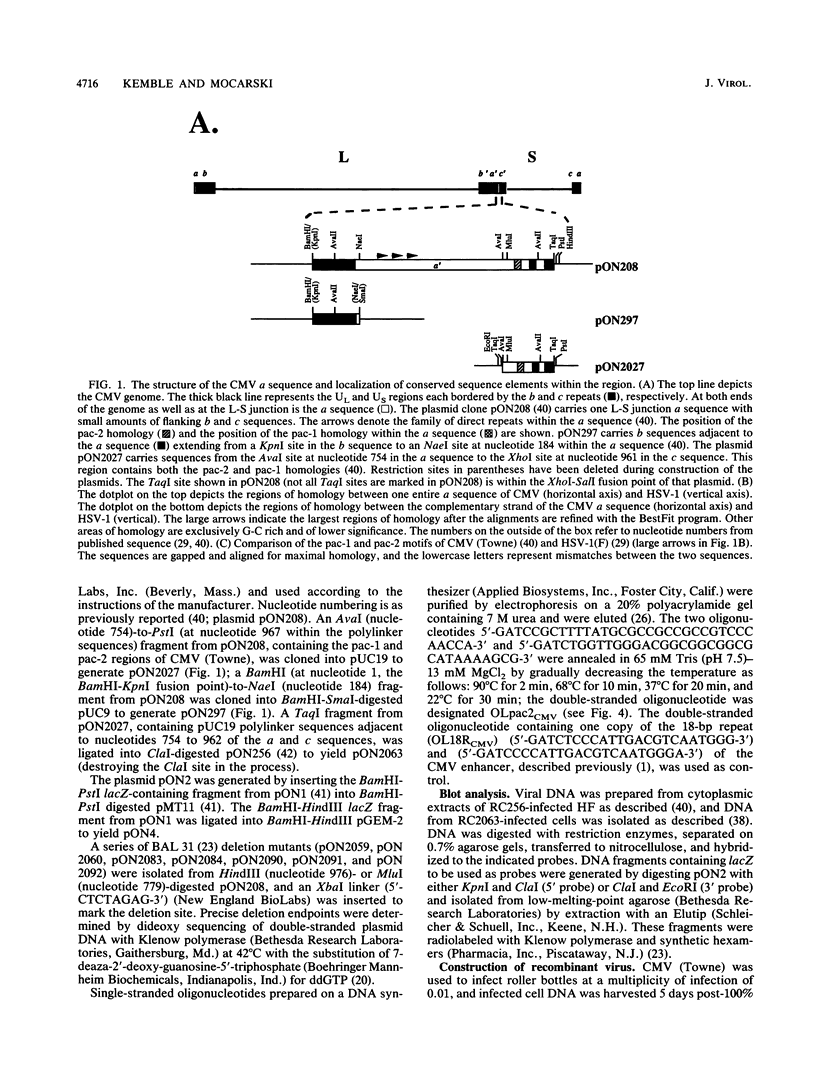
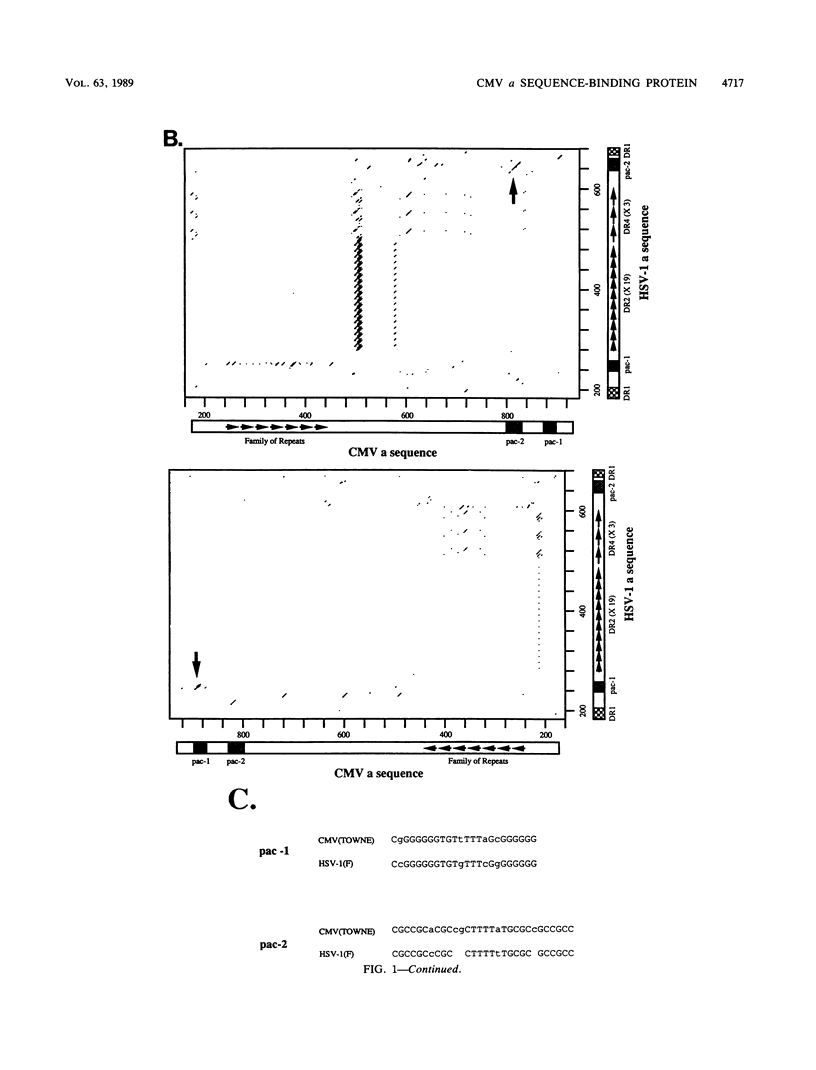
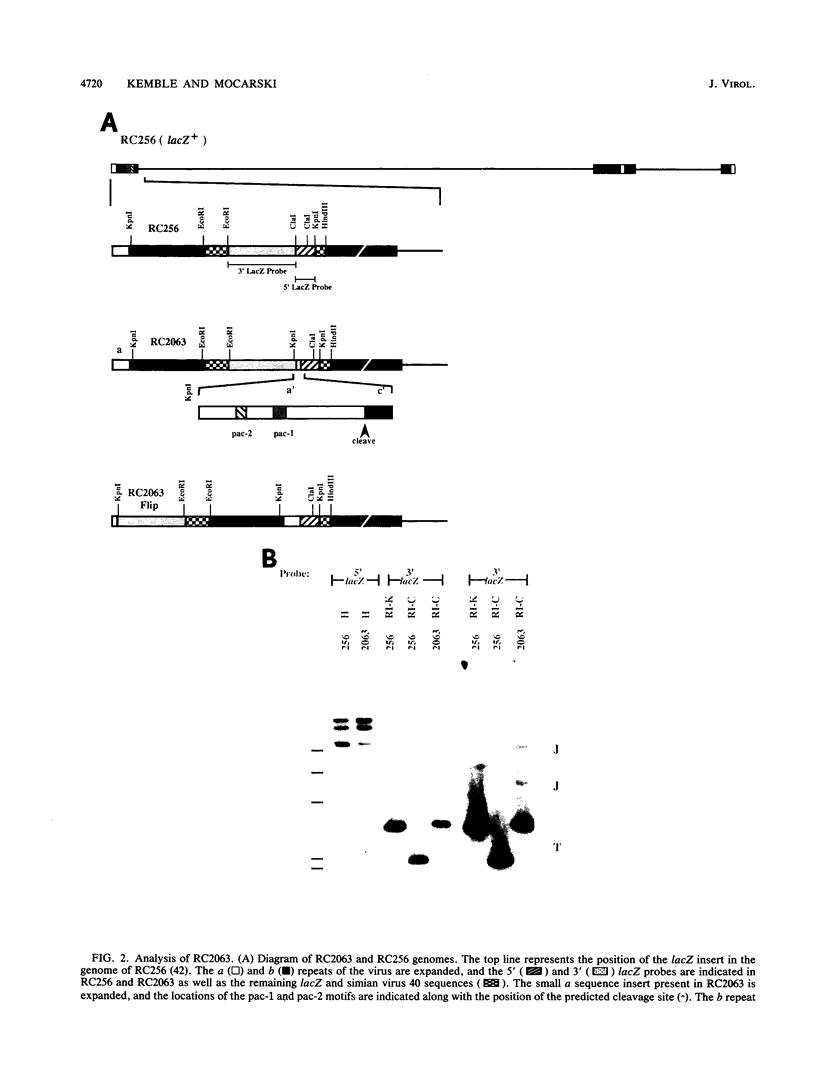
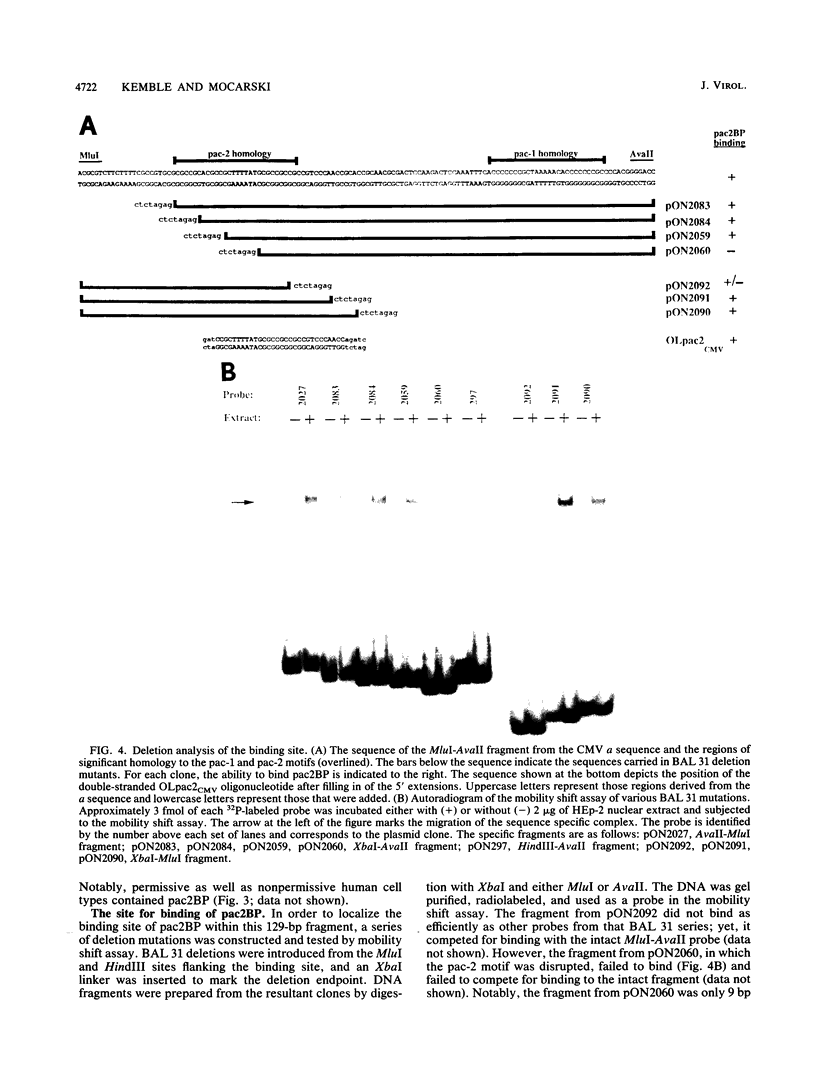
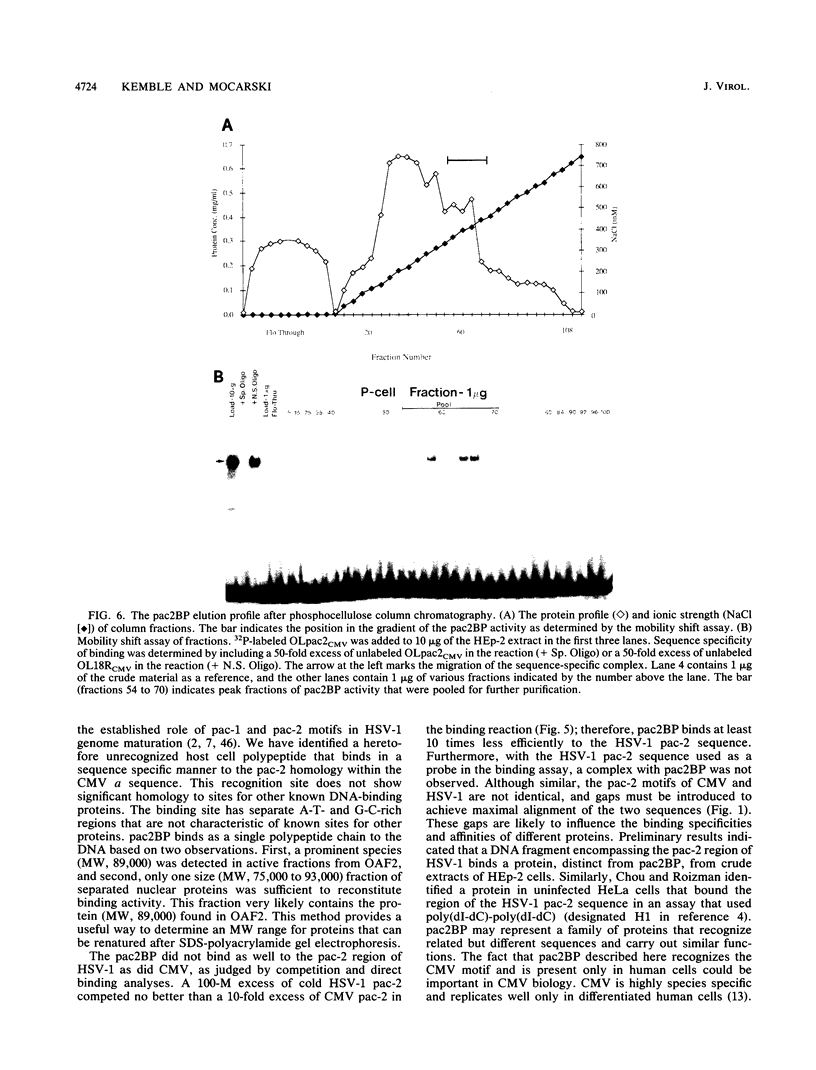
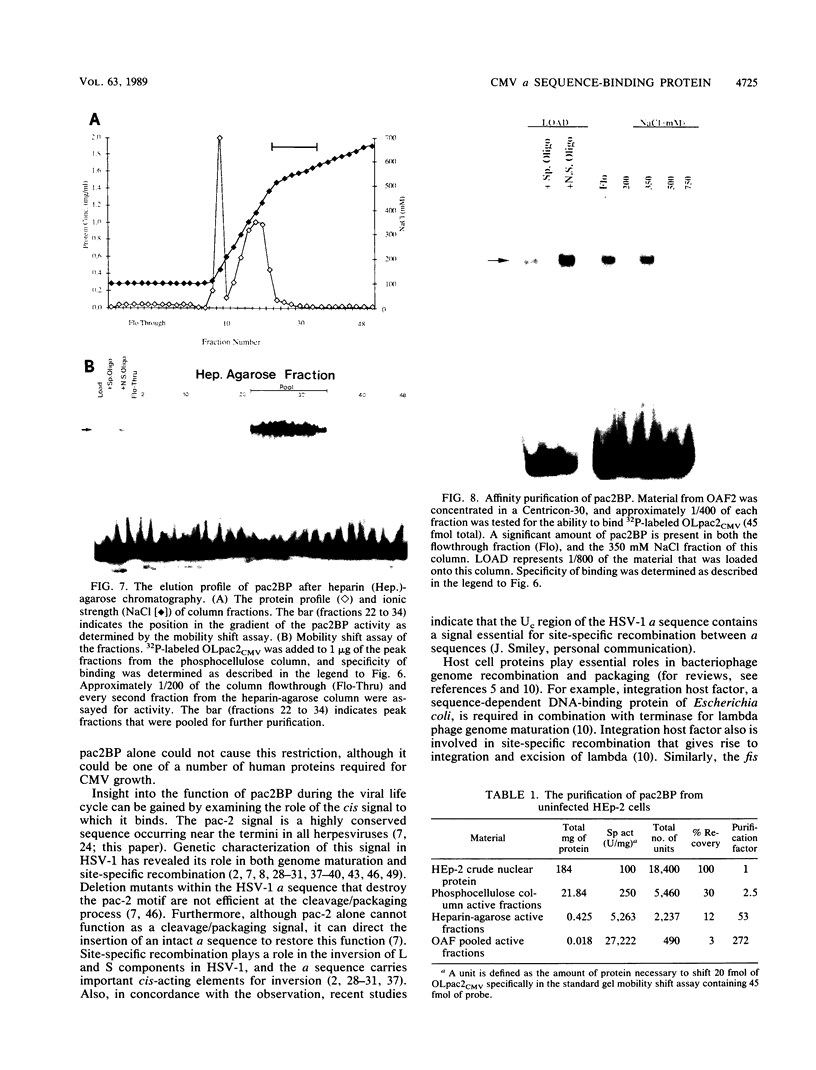
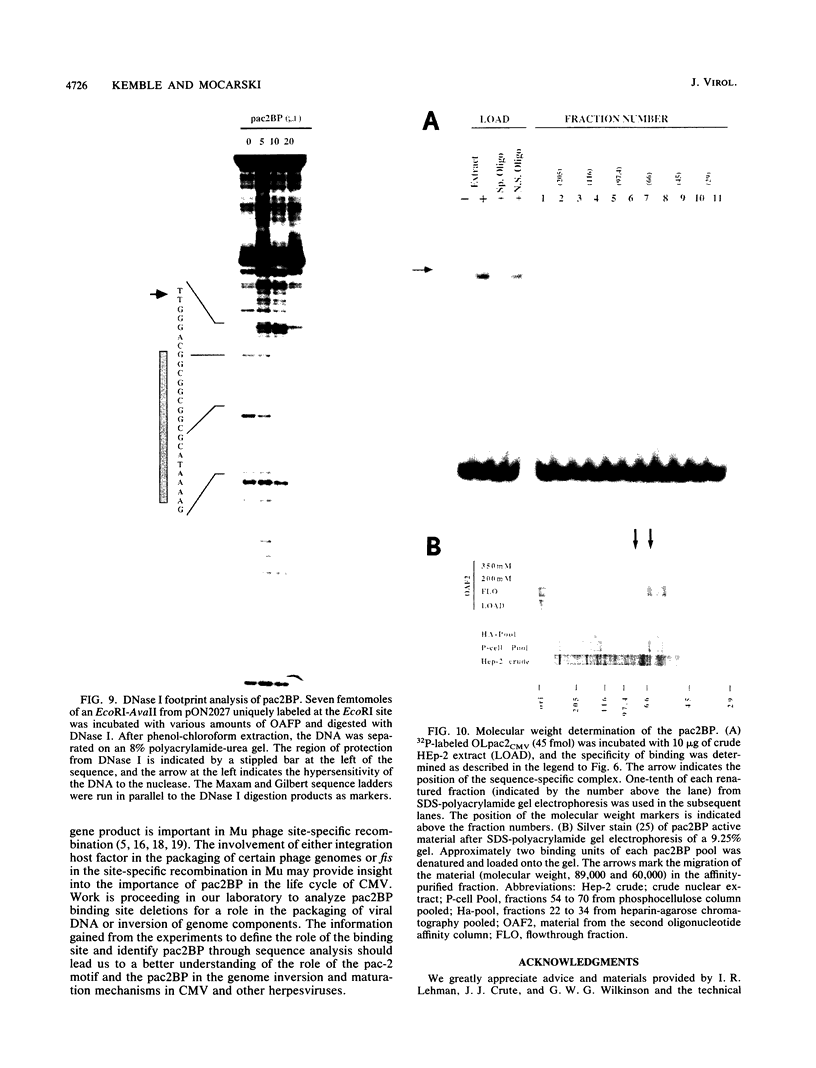
The human cytomegalovirus (CMV) a sequence has significant homology to two regions, pac-1 and pac-2, within the a sequence of herpes simplex virus type 1 (HSV-1). Both regions have been shown to be important cis-acting signals in HSV-1 genome maturation. We have demonstrated that a small fragment from within the CMV a sequence, containing the pac-1 and pac-2 motifs, carries all of the signals necessary for generation of genomic termini and for inversion. These observations indicated that the function of these highly conserved sequence motifs was similar in CMV and HSV-1. We have identified and partially purified a host cell protein with affinity for the sequence 5'-GGCGGCGGCGCATAAAA-3' within CMV pac-2. This partially purified protein has an apparent molecular weight of 89,000 under denaturing conditions and could be renatured after sodium dodecyl sulfate-polyacrylamide gel electrophoresis, suggesting that the capacity to bind DNA was the property of a single polypeptide chain. This activity was found in a wide variety of human cell lines, including those that are permissive as well as those that are nonpermissive for CMV growth, but not in cell lines from monkey, mouse, or drosophila origins. Our work implicates a host cell protein in a sequence function.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cherrington J. M., Mocarski E. S. Human cytomegalovirus ie1 transactivates the alpha promoter-enhancer via an 18-base-pair repeat element. J Virol. 1989 Mar;63(3):1435–1440. doi: 10.1128/jvi.63.3.1435-1440.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J., Roizman B. Characterization of DNA sequence-common and sequence-specific proteins binding to cis-acting sites for cleavage of the terminal a sequence of the herpes simplex virus 1 genome. J Virol. 1989 Mar;63(3):1059–1068. doi: 10.1128/jvi.63.3.1059-1068.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J., Roizman B. Isomerization of herpes simplex virus 1 genome: identification of the cis-acting and recombination sites within the domain of the a sequence. Cell. 1985 Jul;41(3):803–811. doi: 10.1016/s0092-8674(85)80061-1. [DOI] [PubMed] [Google Scholar]
- Chou J., Roizman B. The terminal a sequence of the herpes simplex virus genome contains the promoter of a gene located in the repeat sequences of the L component. J Virol. 1986 Feb;57(2):629–637. doi: 10.1128/jvi.57.2.629-637.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig N. L. Site-specific inversion: enhancers, recombination proteins, and mechanism. Cell. 1985 Jul;41(3):649–650. doi: 10.1016/s0092-8674(85)80040-4. [DOI] [PubMed] [Google Scholar]
- Dalziel R. G., Marsden H. S. Identification of two herpes simplex virus type 1-induced proteins (21K and 22K) which interact specifically with the a sequence of herpes simplex virus DNA. J Gen Virol. 1984 Sep;65(Pt 9):1467–1475. doi: 10.1099/0022-1317-65-9-1467. [DOI] [PubMed] [Google Scholar]
- Deiss L. P., Chou J., Frenkel N. Functional domains within the a sequence involved in the cleavage-packaging of herpes simplex virus DNA. J Virol. 1986 Sep;59(3):605–618. doi: 10.1128/jvi.59.3.605-618.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiss L. P., Frenkel N. Herpes simplex virus amplicon: cleavage of concatemeric DNA is linked to packaging and involves amplification of the terminally reiterated a sequence. J Virol. 1986 Mar;57(3):933–941. doi: 10.1128/jvi.57.3.933-941.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman D. I. Integration host factor: a protein for all reasons. Cell. 1988 Nov 18;55(4):545–554. doi: 10.1016/0092-8674(88)90213-9. [DOI] [PubMed] [Google Scholar]
- Fung M. C., Chiu K. Y., Weber T., Chang T. W., Chang N. T. Detection and purification of a recombinant human B lymphotropic virus (HHV-6) in the baculovirus expression system by limiting dilution and DNA dot-blot hybridization. J Virol Methods. 1988 Jan;19(1):33–42. doi: 10.1016/0166-0934(88)90005-5. [DOI] [PubMed] [Google Scholar]
- Hager D. A., Burgess R. R. Elution of proteins from sodium dodecyl sulfate-polyacrylamide gels, removal of sodium dodecyl sulfate, and renaturation of enzymatic activity: results with sigma subunit of Escherichia coli RNA polymerase, wheat germ DNA topoisomerase, and other enzymes. Anal Biochem. 1980 Nov 15;109(1):76–86. doi: 10.1016/0003-2697(80)90013-5. [DOI] [PubMed] [Google Scholar]
- Jacob R. J., Morse L. S., Roizman B. Anatomy of herpes simplex virus DNA. XII. Accumulation of head-to-tail concatemers in nuclei of infected cells and their role in the generation of the four isomeric arrangements of viral DNA. J Virol. 1979 Feb;29(2):448–457. doi: 10.1128/jvi.29.2.448-457.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean J. H., Yoshimura N., Furukawa T., Plotkin S. A. Intracellular forms of the parental human cytomegalovirus genome at early stages of the infective process. Virology. 1978 May 1;86(1):281–286. doi: 10.1016/0042-6822(78)90029-6. [DOI] [PubMed] [Google Scholar]
- Johnson R. C., Bruist M. F., Simon M. I. Host protein requirements for in vitro site-specific DNA inversion. Cell. 1986 Aug 15;46(4):531–539. doi: 10.1016/0092-8674(86)90878-0. [DOI] [PubMed] [Google Scholar]
- Kadonaga J. T., Tjian R. Affinity purification of sequence-specific DNA binding proteins. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahmann R., Rudt F., Koch C., Mertens G. G inversion in bacteriophage Mu DNA is stimulated by a site within the invertase gene and a host factor. Cell. 1985 Jul;41(3):771–780. doi: 10.1016/s0092-8674(85)80058-1. [DOI] [PubMed] [Google Scholar]
- Koch C., Vandekerckhove J., Kahmann R. Escherichia coli host factor for site-specific DNA inversion: cloning and characterization of the fis gene. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4237–4241. doi: 10.1073/pnas.85.12.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneluk R. G., Quan F., Gravel R. A. Rapid and reliable dideoxy sequencing of double-stranded DNA. Gene. 1985;40(2-3):317–323. doi: 10.1016/0378-1119(85)90055-1. [DOI] [PubMed] [Google Scholar]
- LaFemina R. L., Hayward G. S. Replicative forms of human cytomegalovirus DNA with joined termini are found in permissively infected human cells but not in non-permissive Balb/c-3T3 mouse cells. J Gen Virol. 1983 Feb;64(Pt 2):373–389. doi: 10.1099/0022-1317-64-2-373. [DOI] [PubMed] [Google Scholar]
- Marks J. R., Spector D. H. Replication of the murine cytomegalovirus genome: structure and role of the termini in the generation and cleavage of concatenates. Virology. 1988 Jan;162(1):98–107. doi: 10.1016/0042-6822(88)90398-4. [DOI] [PubMed] [Google Scholar]
- Marshall T. Detection of protein in polyacrylamide gels using an improved silver stain. Anal Biochem. 1984 Feb;136(2):340–346. doi: 10.1016/0003-2697(84)90227-6. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mocarski E. S., Deiss L. P., Frenkel N. Nucleotide sequence and structural features of a novel US-a junction present in a defective herpes simplex virus genome. J Virol. 1985 Jul;55(1):140–146. doi: 10.1128/jvi.55.1.140-146.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Post L. E., Roizman B. Molecular engineering of the herpes simplex virus genome: insertion of a second L-S junction into the genome causes additional genome inversions. Cell. 1980 Nov;22(1 Pt 1):243–255. doi: 10.1016/0092-8674(80)90172-5. [DOI] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Herpesvirus-dependent amplification and inversion of cell-associated viral thymidine kinase gene flanked by viral a sequences and linked to an origin of viral DNA replication. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5626–5630. doi: 10.1073/pnas.79.18.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Site-specific inversion sequence of the herpes simplex virus genome: domain and structural features. Proc Natl Acad Sci U S A. 1981 Nov;78(11):7047–7051. doi: 10.1073/pnas.78.11.7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Structure and role of the herpes simplex virus DNA termini in inversion, circularization and generation of virion DNA. Cell. 1982 Nov;31(1):89–97. doi: 10.1016/0092-8674(82)90408-1. [DOI] [PubMed] [Google Scholar]
- Morse L. S., Pereira L., Roizman B., Schaffer P. A. Anatomy of herpes simplex virus (HSV) DNA. X. Mapping of viral genes by analysis of polypeptides and functions specified by HSV-1 X HSV-2 recombinants. J Virol. 1978 May;26(2):389–410. doi: 10.1128/jvi.26.2.389-410.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasseri M., Mocarski E. S. The cleavage recognition signal is contained within sequences surrounding an a-a junction in herpes simplex virus DNA. Virology. 1988 Nov;167(1):25–30. doi: 10.1016/0042-6822(88)90050-5. [DOI] [PubMed] [Google Scholar]
- Ohlsson H., Edlund T. Sequence-specific interactions of nuclear factors with the insulin gene enhancer. Cell. 1986 Apr 11;45(1):35–44. doi: 10.1016/0092-8674(86)90535-0. [DOI] [PubMed] [Google Scholar]
- Roizman B., Carmichael L. E., Deinhardt F., de-The G., Nahmias A. J., Plowright W., Rapp F., Sheldrick P., Takahashi M., Wolf K. Herpesviridae. Definition, provisional nomenclature, and taxonomy. The Herpesvirus Study Group, the International Committee on Taxonomy of Viruses. Intervirology. 1981;16(4):201–217. doi: 10.1159/000149269. [DOI] [PubMed] [Google Scholar]
- Roizman B. Genome variation and evolution among herpes viruses. Ann N Y Acad Sci. 1980;354:472–483. doi: 10.1111/j.1749-6632.1980.tb27986.x. [DOI] [PubMed] [Google Scholar]
- Smiley J. R., Fong B. S., Leung W. C. Construction of a double-jointed herpes simplex viral DNA molecule: inverted repeats are required for segment inversion, and direct repeats promote deletions. Virology. 1981 Aug;113(1):345–362. doi: 10.1016/0042-6822(81)90161-6. [DOI] [PubMed] [Google Scholar]
- Spaete R. R., Frenkel N. The herpes simplex virus amplicon: a new eucaryotic defective-virus cloning-amplifying vector. Cell. 1982 Aug;30(1):295–304. doi: 10.1016/0092-8674(82)90035-6. [DOI] [PubMed] [Google Scholar]
- Spaete R. R., Frenkel N. The herpes simplex virus amplicon: analyses of cis-acting replication functions. Proc Natl Acad Sci U S A. 1985 Feb;82(3):694–698. doi: 10.1073/pnas.82.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaete R. R., Mocarski E. S. Insertion and deletion mutagenesis of the human cytomegalovirus genome. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7213–7217. doi: 10.1073/pnas.84.20.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaete R. R., Mocarski E. S. Regulation of cytomegalovirus gene expression: alpha and beta promoters are trans activated by viral functions in permissive human fibroblasts. J Virol. 1985 Oct;56(1):135–143. doi: 10.1128/jvi.56.1.135-143.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaete R. R., Mocarski E. S. The alpha sequence of the cytomegalovirus genome functions as a cleavage/packaging signal for herpes simplex virus defective genomes. J Virol. 1985 Jun;54(3):817–824. doi: 10.1128/jvi.54.3.817-824.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow N. D., McMonagle E. C., Davison A. J. Fragments from both termini of the herpes simplex virus type 1 genome contain signals required for the encapsidation of viral DNA. Nucleic Acids Res. 1983 Dec 10;11(23):8205–8220. doi: 10.1093/nar/11.23.8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss F., Varshavsky A. A protein binds to a satellite DNA repeat at three specific sites that would be brought into mutual proximity by DNA folding in the nucleosome. Cell. 1984 Jul;37(3):889–901. doi: 10.1016/0092-8674(84)90424-0. [DOI] [PubMed] [Google Scholar]
- Tamashiro J. C., Spector D. H. Terminal structure and heterogeneity in human cytomegalovirus strain AD169. J Virol. 1986 Sep;59(3):591–604. doi: 10.1128/jvi.59.3.591-604.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmuza S. L., Smiley J. R. Signals for site-specific cleavage of HSV DNA: maturation involves two separate cleavage events at sites distal to the recognition sequences. Cell. 1985 Jul;41(3):793–802. doi: 10.1016/s0092-8674(85)80060-x. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Villarreal L. P., Berg P. Hybridization in situ of SV40 plaques: detection of recombinant SV40 virus carrying specific sequences of nonviral DNA. Science. 1977 Apr 8;196(4286):183–185. doi: 10.1126/science.191907. [DOI] [PubMed] [Google Scholar]
- Vlazny D. A., Kwong A., Frenkel N. Site-specific cleavage/packaging of herpes simplex virus DNA and the selective maturation of nucleocapsids containing full-length viral DNA. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1423–1427. doi: 10.1073/pnas.79.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidekamm E., Wallach D. F., Flückiger R. A new sensitive, rapid fluorescence technique for the determination of proteins in gel electrophoresis and in solution. Anal Biochem. 1973 Jul;54(1):102–114. doi: 10.1016/0003-2697(73)90252-2. [DOI] [PubMed] [Google Scholar]