Figure 2. Time behavior of the msd of the six inner beads (g
1(t), continuous lines), compared to the average square gyration radius 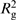 (horizontal dashed lines) of the whole chromosome and measurements of the msd of the active GAL gene inside in vivo yeast nuclei [24] (purple dots).
(horizontal dashed lines) of the whole chromosome and measurements of the msd of the active GAL gene inside in vivo yeast nuclei [24] (purple dots).
For comparison, g 1(t) for yeast chromosomes without topological constraints has been shown (cyan line). On short time scales, our model reproduces the typical dynamics of semi-flexible polymers with g 1(t)∼t 0.75 [25]. For the model with constraints, there is no extended Rouse regime due to the insufficient separation of Kuhn and entanglement length. Nevertheless, we observe the characteristic g 1(t)∼t 0.25 regime for entangled, flexible polymers [7].

