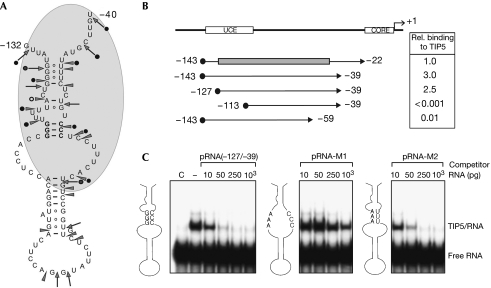
Figure 2.
TIP5 recognizes the secondary structure of promoter RNA. (A) Schematic depicting the secondary structure of mouse pRNA as shown by RNase footprinting. Cleavages by RNase T1 are indicated by arrows, and RNase V1 cleavages by arrowheads. Cleavages that are decreased as a consequence of human TIP5 binding are indicated by black circles. The area of pRNA that is shielded from RNase cleavage is shaded in grey. The three G-C base pairs mutated in pRNA-M1 are indicated in bold. (B) Mapping of the minimal TIP5 interacting RNA domain. Black circles indicate T7 promoter-derived sequences common to all run-off transcripts used. The grey box marks the part of pRNA that is involved in hairpin formation. The numbers on the right are taken from Table 1, representing the relative binding affinities of murine TIP51−598. (C) Mutations that disrupt the secondary structure abrogate TIP5 binding to pRNA. Mouse pRNA and mutants pRNA-M1 and pRNA-M2 comprising the minimal sequence required for TIP5 binding (from −127 to −39) were used in EMSA competition assays containing murine TIP51−598 and labelled MCS-RNA. The schemes indicate nucleotide exchanges in pRNA-M1 and -M2 and the predicted structure of the individual pRNAs. pRNA, promoter RNA.