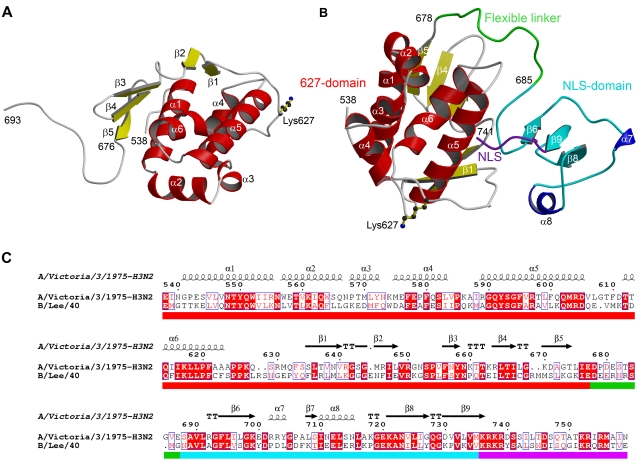
Figure 1. Structure and sequence alignment of C-terminal domains of influenza polymerase PB2 subunit.
(A) Ribbon diagram of the 627-domain showing secondary structure elements and the position of human specific lysine 627. Helices are in red and beta-strands in yellow as defined by DSSP [30]. The conformation of the C-terminal tail (residues 676–693) is determined by crystal contacts. The structure shown is of the SeMet labelled protein. (B) Ribbon diagram of the 627-NLS-double domain showing the position of lysine 627. The 627-domain is in red and yellow, the core NLS-domain in cyan and blue and the truncated nuclear localization peptide in purple. The flexible inter-domain linker is in green. Figure 1A and Figure 1B were drawn with MOLSCRIPT [42] and rendered with RASTER3D [43]. (C) Sequence alignment of C-terminal regions of PB2 from influenza A and B viruses with superimposed secondary structure. The coloured bar under the alignment indicates the 627-domain (red), linker (green), core NLS-domain (cyan) and the bipartite NLS (purple). The seven host specific residues identified in this region [25] are indicated with a blue square in the coloured bar. Alignment figure produced with ESPript [44].