Abstract
The Darkener of apricot (Doa) locus of Drosophila encodes a LAMMER protein kinase affecting alterative splicing, and hence sex determination, via the phosphorylation of SR and SR-like proteins. Doa encodes 6 different kinases via alternative promoter usage. To provide further insight into the roles of the multiple isoforms, we mapped polymorphisms, deletions, and P-element insertions in the locus, identifying several that are largely, if not completely, isoform specific in their effects. These tests, along with the use of lines permitting overexpression and interfering RNA expression, demonstrate that the major isoforms of 55 and 105 kDa perform separate functions. The 105-kDa and a minor 138-kDa isoform are both vital but do not apparently perform functions essential for sex determination. Curiously, male-specific lethality induced by overexpression of the 55-kDa kinase in the larval fat body is rescued by coexpression of TRA, suggesting a sex-specific physiological role for this isoform. Maternal effects in which the survival of heteroallelic adults depends upon the direction of the cross are consistent with a role for a 105-kDa cytoplasmic kinase in oogenesis or early larval development.
THE Darkener of apricot (Doa) locus of Drosophila encodes a protein kinase of the LAMMER or Clk family (Yun et al. 1994). Members of this kinase family phosphorylate their substrates on Ser residues, although they also autophosphorylate on Thr and Tyr (Ben-David et al. 1991; Howell et al. 1991; Lee et al. 1996; Nayler et al. 1997). SR and SR-“like” proteins are substrates both in vitro and in vivo for LAMMER kinases from diverse organisms, including mammals, Drosophila, and plants (Colwill et al. 1996; Nayler et al. 1997; Du et al. 1998; Savaldi-Goldstein et al. 2000). A consensus phosphorylation site determined for several LAMMER family members confirms this specificity, but also suggests that specificity determinants exist in addition to the simple repeating motif “RSRSRS” associated with SR and SR-like proteins (Nikolakaki et al. 2002).
Doa mutations suppress phenotypes due to copia retrotransposon insertions at three loci and are almost invariably recessive lethal (Rabinow and Birchler 1989; Rabinow et al. 1993). Rare heteroallelic survivors of specific genotypes reveal characteristic phenotypes, which depend upon the alleles involved. Degeneration of retinal photoreceptors is observed in rare homozygotes of two alleles associated with position-effect variegation (Yun et al. 1994). A weak Doa allele was also isolated due to its effects of increasing bristle number in a screen for QTL loci (Norga et al. 2003).
The analysis of both somatic and germ-line cell clones demonstrated that cells homozygous for null alleles of Doa are inviable (Yun et al. 2000). Consistent with the necessity of Doa function in the germ line, two alleles were also recovered in a clonal mutagenic screen for loci influencing early oocyte development (Morris et al. 2003).
Although homozygous Doa larvae derived from heterozygous mothers hatch normally, they die during early larval stages, suggesting that maternal contribution of Doa activity rescues homozygous mutants during early development. This interpretation is supported by the observation that homozygous embryos derived from heteroallelic mothers show variable defects, including the disruption of differentiation in the ventral nervous system and loss of normal cuticular segmentation (Yun et al. 1994).
Doa function has been implicated by genomic screens in a number of cellular, differentiative, and behavioral processes, including progression through G1 phase of the cell cycle (Bettencourt-Dias et al. 2004; Bjorklund et al. 2006), autophagy (Gorski et al. 2003; Lee et al. 2003), and protein secretion (Bard et al. 2006). Mutant alleles of the kinase were also recently isolated in screens for increased aggression (Dierick and Greenspan 2006; Edwards et al. 2006).
The best characterized molecular function of DOA kinase and its orthologs in other species is the regulation of alternative splicing via the direct phosphorylation of SR proteins (Colwill et al. 1996; Nayler et al. 1997; Du et al. 1998; Prasad et al. 1999; Savaldi-Goldstein et al. 2000; Nikolakaki et al. 2002). In Drosophila, SR and SR-like proteins such as TRA and TRA2 regulate somatic sex determination (Saccone et al. 2002; Forch and Valcarcel 2003). The phosphorylation of SR and SR-like proteins influences their interactions with each other as well as with their RNA substrates. We previously demonstrated that Doa alleles induce mild female-to-male somatic sexual transformations, hypophosphorylation, and mislocalization of some of the SR and SR-like proteins and altered splicing of the dsx transcript, encoding a key sex-regulatory protein (Du et al. 1998). Furthermore, RNAi “knockdown” of Doa influences some, but not all splicing events in cultured cells (Park et al. 2004).
Doa expresses at least six different isoforms, which are expressed with developmental specificity from alternative promoters (Kpebe and Rabinow 2008). Particularly high levels of kinase-encoding transcripts, and the use of an alternative polyadenylation site rendering some of the transcripts unstable during early embryogenesis, suggest that a high level of kinase activity is required during the activation of zygotic gene expression.
The multiple DOA isoforms possess virtually identical kinase catalytic domains but different noncatalytic N termini. High levels of amino acid conservation in the catalytic domains of LAMMER kinases are generally observed, for example, 75% amino acid identity between Drosophila DOA and human CLK2. However, sequence comparisons outside the catalytic domains break down over larger phylogenetic distances, suggesting that the sequence of the N-terminal noncatalytic domains is not essential to their activity. Nevertheless, alignments between the alternative N-terminal exons of Drosophila melanogaster Doa and the genomic sequences of other Drosophila species revealed that the N-terminal noncatalytic exons of the kinase are conserved in all 11 species examined, e.g., in excess of 50% amino acid identity between D. melanogaster and D. virilis, which are separated by ∼60 million years of evolutionary divergence (Kpebe and Rabinow 2008). This observation suggests that the divergent N termini may play important roles in DOA function.
Given these observations, as well as the pleiotropic nature of Doa phenotypes and the complexity of genetic interactions among the previously tested alleles, we wished to clarify the functions of the multiple proteins encoded at the locus. Here we describe alleles, which are primarily specific for one or two isoforms. Some functions of the multiple isoforms are independent or may even oppose each other, while others are overlapping. Partial complementation of two kinase-null alleles suggests that the multiple alternative noncatalytic N termini provide some necessary function. Finally, overexpression of one of the kinase isoforms in the larval fat body results in male-specific lethality, which is rescued by simultaneous coexpression of TRA-F, suggesting that the kinase alters essential male-specific physiology of this organ.
MATERIALS AND METHODS
Drosophila stocks, crosses, and mutageneses:
D. melanogaster were raised on cornmeal medium at 25°. Crosses to test for phenotypes using GAL4-directed expression were performed at 29° to enhance phenotypes. Many Doa mutant strains and heteroallelic combinations were previously described (Rabinow et al. 1993; Yun et al. 1994). Five previously unpublished Doa alleles were generous gifts from James Birchler. Four of these (γ3A, γ3C, γ3E, and γ3G), were induced by irradiation with 4000 rad from a 137Cs source. Progeny were screened for suppression of whiteapricot. The fifth allele, EMS-C, is homozygous viable, EMS induced, and also recovered from progeny screened for suppression of wa. Recombination mapping placed EMS-C at 3-99, the location of Doa. The Doa allele Su(Mrt)9, a gift from Brad Phillips, was isolated during screens for suppressors of the hypermorphic hh allele, Mrt. It is likely due to a chromosomal rearrangement on 3R. The Δ2-3 P element was used to mobilize the l(3)s2784 P element to generate precise excisions. w− progeny were recovered from a cross of heterozygotes for the Δ2-3 chromosome and the l(3)s2784 P element. These were screened for lethality in combination with Df(3R)3450. The activating insertion alleles P{EP}DoaEP3080, P{EPgy2}DoaEY11301, and P{EPgy2}DoaEY08857 are described in FlyBase (http://flybase.org/) and were obtained from the Bloomington Stock Center. The insertion sites of all P-element alleles were verified by inverse PCR. GAL4 lines (Brand and Perrimon 1993) were obtained from Bloomington and Jean-René Martin.
Transgenic lines expressing Doa isoforms and interfering RNAs:
Two lines producing interfering RNA (RNAi) specific for the 105-kDa coding transcript (“RE”, CG31049) were very generously provided prior to release, by Ryu Ueda and colleagues at the National Institute of Genetics (NIG), Genetic Strains Research Center. In these lines, a genomic fragment of ∼500 bp corresponding to an exon present in the 105-kDa Doa isoform was generated with the primers 5′-aaggcctacatggccggaccgACAGCGTTAGCAGCAGCAACA and 3′-aatctagaggtaccTACGGAGCCTGCTGGCTGATA.
A single line expressing an interfering RNA for CG33204, the 138-kDa DOA isoform, was obtained from the Vienna Drosophila RNAi stock center (VDRC) (Dietzl et al. 2007). Additionally, a complete cDNA of the 69-kDa DOA isoform from EST RE21871, including the full-length 3′-UTR, was cloned into the pUAS-T vector (Kpebe and Rabinow 2008). A second transgene, using a shortened 3′-UTR expressed only during early embryonic development, was similarly constructed. w1118 embryos were injected by the Duke University Non-Mammalian Model Systems Flyshop. A minimum of five lines were recovered and analyzed for each construct.
Histology and scanning electron microscopy:
For semithin retinal sections, fly heads were dissected and fixed in 2% glutaraldehyde in 0.1 m Na-cacodylate (pH 7.3) buffer and then in 1% OsO4 in cacodylate buffer, after which they were dehydrated in an ascending alcohol series (30, 50, 70, 90%). They were then put through an ascending series of 2-hydroxypropylmethacrylate (HMPA) and 5% EtOH, 95% HMPA; 3% EtOH, 97% HMPA—EPON:HMPA, 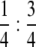 ; EPON:HMPA,
; EPON:HMPA, 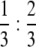 ; EPON:HMPA, 1:1; and benzyl dimethylamine (BDMA). Sections of 1 μm were cut on a Leica Ultracut UCT ultramicrotome and stained with a mixture of 1:1 1% methylene blue in 1% borax and 1% Azur V.
; EPON:HMPA, 1:1; and benzyl dimethylamine (BDMA). Sections of 1 μm were cut on a Leica Ultracut UCT ultramicrotome and stained with a mixture of 1:1 1% methylene blue in 1% borax and 1% Azur V.
For scanning electron micrography, flies were dehydrated through an ascending ethanol series (30, 50, and 70%), slowly frozen at −18° under partial vacuum (90 Pa) on a Peltier stage, and observed under ESED mode on a Hitachi S3000N scanning electron microscope (acceleration voltage 10–12 kV).
In situ hybridization to whole embryos was carried out as described (Tautz and Pfeifle 1989), using digoxigenin labeling and detection (Roche, Indianapolis). Staging of embryos was as per Campos-Ortega and Hartenstein (1997).
Molecular biology:
Inverse PCR was performed following the Berkeley Drosophila Genome Project protocol (http://www.fruitfly.org/). For single-stranded conformational polymorphism (SSCP), as performed by (Villard et al. 2000), the coding region and the 3′-UTR of the Doa locus were subdivided into 29 fragments of ∼300 bp in length. PCR reactions were performed in a final volume of 50 μl, using 15 pmol of primer and 100 ng of Drosophila genomic DNA. Primers were designed from genomic sequences for annealing at 60° (supplemental Table 1). Amplification was for 30 cycles consisting of 45 sec denaturation at 94°, 45 sec annealing, and 45 sec extension at 72°. The PCR reaction was mixed with an equal volume of freshly prepared 0.1 n NaOH. Following denaturation for 4 min at 94°, samples were cooled on ice and loaded onto a 6% polyacrylamide gel. Electrophoresis was carried out with a constant current of 10 mA twice, once at room temperature and once at 4°, to reveal the maximum number of sequence alterations. Polymorphic sites were confirmed by DNA sequencing.
RNA preparations, northern transfers, and probe preparation were performed as described (Kpebe and Rabinow 2008). Loading controls were performed by rehybridizing the transfers with a probe for rp49 (O'Connell and Rosbash 1984).
RESULTS
Molecular analysis of Doa alleles:
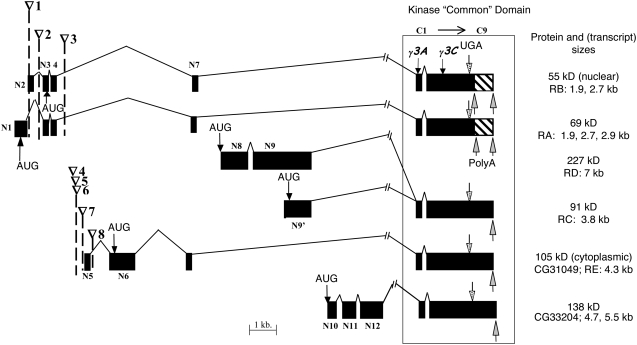
Six alternative isoforms of DOA kinase possess different N-terminal noncatalytic domains and a virtually identical C-terminal “common” domain (Figure 1) (Kpebe and Rabinow 2008). The common domain encompasses the kinase catalytic domain and one additional N-terminal exon present in all isoforms. One obvious question concerning these isoforms is whether they have identical, overlapping, or completely independent functions, which could be elucidated by molecular analysis of complementing alleles.
Figure 1.—
Schematic of the structure of six verified and predicted DOA isoforms. Exons are drawn as solid boxes, each molecular weight designating a different isoform. The corresponding BDGP transcript designations are also shown (“RA” to “RE.”), as are their verified sizes. Exons are defined and numbered as either in the N-terminal variable region (N1–12) or in the region common to all the kinase isoforms (C1–9; box), which includes the kinase catalytic domain. The nine exons in the common domain are too small to be indicated. All transposon insertions shown are P elements with the exception of the HD allele, which is a copia retrotransposon. ∇1, DoaEP3080; ∇2, DoaEY11301; ∇3, Doa HD; ∇4, l(3)04743; ∇5, l(3)01705; ∇6, l(3)s2784; ∇7, EY08857; ∇8, l(3)S084402. The precise locations of the eight transposon insertions and the two small deletions are presented in Table 1. Molecular weights of the proteins are either from experimental evidence (55, 69, 91, and 105 kDa), or predicted from BDGP sequence analyses for the two nonverified protein isoforms (138 kDa, 227 kDa). Translation start and stop sites are indicated (AUG, UGA). An alternative polyadenylation site producing 55- and 69-kDa coding transcripts with truncated 3′-UTRs during the first 4 hr of embryogenesis is indicated by an upward arrow; hatch marks indicate the region eliminated in these transcripts.
With two exceptions, SSCP analysis (nine alleles) and sequencing of virtually all Doa exons and the 3′-UTR (four alleles) failed to reveal any sequence alterations that could cause Doa mutations (supplemental Table 2; see legend for additional details). Changes identified in sequences outside the catalytic domain and in the 3′-UTR include single-nucleotide polymorphisms, insertions, and deletions of between one and three amino acids. In contrast, no amino acid substitutions were observed in the catalytic domain. Sequence alterations identified in the catalytic domain lie in introns, induce silent changes, or in two cases cause mutations.
Both mutations identified lie in the catalytic domain. Doaγ3A has a 14-nucleotide deletion (accession EF489457), and Doaγ3C possesses a 4-nucleotide deletion (accession EF489458). Both deletions cause frameshifts, the introduction of premature stop codons, and the production of C-terminally truncated DOA proteins (Kpebe and Rabinow 2008).
Characterization of transposable element-induced isoform-specific Doa alleles:
A copia insertion specifically affects Doa 2.7-kb transcripts:
The DoaHD allele, isolated during hybrid dysgenic screens for mutations modifying whiteapriot (Rabinow and Birchler 1989; Yun et al. 1994), was caused by a copia retrotransposon inserted anti-parallel to the gene at nucleotide 7791 (reference sequence NG_000621; Figure 1; Table 3). On the basis of the transcription termination effects of copia insertions in other loci (Zachar et al. 1985; Campuzano et al. 1986), this insertion would be expected to reduce accumulation of the 2.7-kb transcripts coding the 55- and 69-kDa proteins. In fact, these transcripts are notably reduced, while the 4.3-kb transcript, originating from a downstream promoter, is largely unaffected in two Doa heteroallelic combinations involving the HD allele (Yun et al. 1994). su(wa), which modifies the phenotypes of some copia and gypsy-induced alleles, had no effect on viability of DoaHD homozygotes (data not shown).
TABLE 3.
Summary of phenotypes observed in Doa heteroallelic combinations
| 1. Eyes | a. Rough, slight ommatidial disorganization |
| b. Mosaic, nonuniform color | |
| c. Grossly disorganized ommatidia | |
| 2. Bristles: scutelar, postalar, dorsocantral, head, or other | a. Duplicated |
| b. Missing | |
| c. Forked | |
| d. Misoriented | |
| 3. Wings | a. Additional or incomplete cross-veins |
| b. Additional or incomplete longitudinal veins | |
| c. “Fragile,” damaged | |
| d. Heavy, blackened veins | |
| e. Held out | |
| f. Weak curly-like phenotype | |
| 4. Sex transformation | a. Female vaginal bristles nonsymmetric, missing |
| b. Male clasper(s) on female vaginal plate | |
| c. Sex comb on female prothoracic legs | |
| d. Male genitals rotated | |
| 5. Aberrant sex ratio in F1 | a. ≥3× female/male F1 |
| b. ≥3× male/female F1 | |
| 6. Deformed legs | a. Metathoracic |
| b. Mesothoracic | |
| 7. Substantial death during pupation | |
| 8. Nonreciprocal results (Table 4) |
A P-element insertion reduces accumulation of the 4.3-kb transcript:
Several putative and proven Doa P-element insertion mutations were obtained from the Bloomington and Szeged stock centers, from the BDGP, and as a gift from Eyal Schejter. The l(3)s2784 insertion was previously localized (Norga et al. 2003), and three additional P-element insertions were identified through complementation tests (Table 1, Figure 1). The insertion sites of three of these alleles were mapped within 900 bp of each other, just preceding the 5′ end of the 4.3-kb transcript encoding a 105-kDa cytoplasmic protein. An additional insertion lies immediately downstream of the first (noncoding) exon of this transcript (Table 1, Figure 1). All four alleles are recessive lethal and fail to complement a deficiency of the region, Df(3R)3450.
TABLE 1.
Summary of lesions, analysis, and origins of Doa alleles
| Allele | SSCP/exons sequenced | Insertion alleles: ∇, Figure 1/position of insertion/deletion relative to NG_000621 | Description of insertion site or mutation | Source |
|---|---|---|---|---|
| DoaDem | Unknown | Rabinow et al. (1993) | ||
| DoaHD | ∇3, Copia element/7791 | 177 nt 3′ to the first noncoding exon (55 kDa; in intron), 231 nt 5′ to the 55-kDa transcript's initiating AUG | Rabinow and Birchler (1989) | |
| Doa105 | Breakpoint outside gene | Rabinow and Birchler (1989) | ||
| DoaEMS-1 | + | Unknown | Rabinow and Birchler (1989) | |
| DoaEMS-2 | + / + | Unknown | Rabinow and Birchler (1989) | |
| DoaEMS-3 | + | Unknown | Rabinow and Bircher (1989) | |
| DoaEMS-C | + / + | Unknown | This report | |
| DoaCC | Breakpoint outside gene | Rabinow and Birchler (1989) | ||
| DoaI5 | + | Unknown | Rabinow and Birchler (1989) | |
| DoaE777 | + | Unknown | Kurkuloset al. (1991) | |
| DoaE786 | + | Unknown | Kurkuloset al. (1991) | |
| DoaREMγA | Breakpoint, PEV-associated allele | Yun et al. (1994); C. So and L. Rabinow (unpublished results) | ||
| DoaMsu1 | Breakpoint, PEV-associated allele | Csink and Birchler (1994); C. So and L. Rabinow (unpublished results) | ||
| DoaMsu2 | Breakpoint, PEV-associated allele | Csink and Birchler (1994); C. So and L. Rabinow (unpublished results) | ||
| Doa12.44 | + / + | Unknown (see text for details) | Morris et al. (2003) | |
| Doa198.14 | + / + | Unknown (see text for details) | Morris et al. (2003) | |
| DoaSu(Mrt9) | Unknown | B. Phillips (personal communication) | ||
| l(3)s2784 | ∇6, P element/8635 | 126 nt 5′ to the first noncoding exon of the 105-kDa coding transcript, 1662 nt 5′ to its initiating AUG | Norga et al. (2003) | |
| l(3)01705 | ∇5, P element/8630 | 131 nt 5′ to the first noncoding exon of the 105-kDa coding transcript, 1667 nt 5′ to its initiating AUG | Spradinget al. (1999); this report | |
| l(3)04743 | ∇4, P element/8106 | 655 nt 5′ to the first noncoding exon of the 105-kDa coding transcript, 2191 nt 5′ to its initiating AUG | Spradinget al. (1999); this report | |
| l(3)S084402 | ∇8, P element/8990 | 229 nt 3′ to the first noncoding exon of the 105-kDa coding transcript, 1307 nt 5′ to its initiating AUG | Deaket al. (1997); this report | |
| Doaγ3A | + | 14-nucleotide deletion/36,392 | Deletion in catalytic domain coding sequence | This report |
| Doaγ3B | Breakpoint within gene | Yun et al. (1994) | ||
| Doaγ3C | + | 4-nt deletion/37,656 | Deletion in catalytic domain coding sequence | This report |
| Doaγ3E | Unknown | This report | ||
| Doaγ3G | Unknown | This report | ||
| P{EP}DoaEP3080 | ∇1, P element/5851 | 45 nt 5′ to the first noncoding exon (55 kDa), 780 nt 5′ to its initiating AUG | Bellenet al. (2004) | |
| P{EPgy2}DoaEY11301 | ∇2, P element/6400 | 177 nt 3′ to the first noncoding exon (55 kDa; in intron), 231 nt 5′ to the 55-kDa transcript's initiating AUG | Bellenet al. (2004) | |
| P{EPgy2}DoaEY08857 | ∇7, P element/8714 | 47 nt 5′ to the first noncoding exon of the 105-kDa coding transcript, 1583 nt 5′ to its initiating AUG | Bellenet al. (2004) |
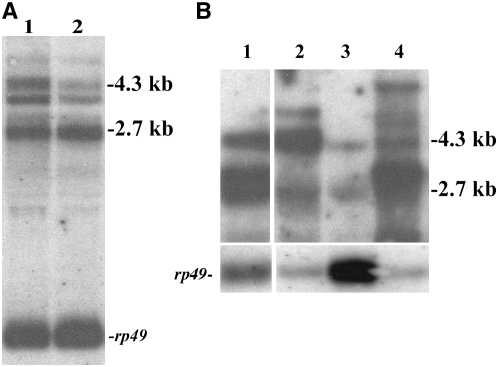
RNA derived from l(3)s2784/DoaHD heteroallelic adult flies shows a marked reduction in the amount of the 4.3-kb transcript compared with levels of the 2.7-kb transcripts (Figure 2A). Since the HD allele reduces levels of the 2.7-kb transcripts without notable effects on the 4.3-kb transcript, we conclude that l(3)s2784 is largely, although perhaps not exclusively isoform specific, reducing transcripts encoding the 105-kDa cytoplasmic DOA protein.
Figure 2.—
Analysis of Doa isoform-specific transcripts in a mutant and UAS-directed expression lines. The northern transfers in both A and B were probed with a 1.8-kb partial cDNA corresponding to the catalytic domain and the N-terminal noncatalytic domain of the 55-kDa coding transcript, which reveals all Doa isoforms (Yun et al. 1994; Kpebe and Rabinow 2008). Transfers were reprobed with rp49 as a loading control. (A) The P element inserted in l(3)s2784 primarily reduces levels of the 4.3-kb Doa transcript encoding the 105-kDa cytoplasmic kinase. Lane 1, wild type (CS) female; lane 2, l(3)s2784/DoaHD female. (B) GAL4-directed expression of specific Doa transcripts via EP and EY P transposons. Northern transfers characterized the isoform specificity of GAL4-directed expression of DoaEP3080, DoaEY11301, and DoaEY08857on RNA from adult heads, probed with the Doa catalytic domain, revealing all isoforms. Exon-specific probes were also used (not shown). Lane 1, GMR-GAL4/+; DoaEY11301/+; lane 2, GMR-GAL4/+; Doa EY08857/+; lane 3, wild type (CS); lane 4, GMR-GAL4/+; DoaEP3080/+. The EP3080 P element directs expression of the entire 55-kDa coding transcript and is homozygous viable, although it will not survive indefinitely without a balancer chromosome present. The EY11301 insertion expresses a transcript lacking the first (noncoding) exon of the 55-kDa coding transcript but including the entire open-reading frame (Figure 1). The P element in EY08857 is inserted immediately 5′ to the start site of the 4.3-kb transcript, which encodes the 105-kDa protein. Note the appearance of a predominant new transcript of slightly higher molecular weight than the normal 2.7-kb RNA (55-kDa coding transcript) in lanes 1 and 4, corresponding to transcripts originating in the inserted EY and EP P elements in these alleles. Amounts of the 4.3-kb transcript encoding the 105-kDa protein are unaffected in DoaEP3080. Levels of a 4.3-kb transcript are increased when the DoaEY11301 P element is activated, but we did not investigate its structure. It may correspond to the native 4.3-kb transcript or may be due to transcripts containing P-element sequences. In contrast, the 4.3-kb (105-kDa coding) transcript is expressed at notably higher levels compared with wild type when driven with GMR-GAL4 in DoaEY08857, while the 55-kDa coding transcript at 2.7 kb remains unaffected (lane 2). A new transcript, slightly >4.3 kb, is also detected in RNA extracts of DoaEY08857, again presumably due to transcription initiation within the inserted P element.
To determine whether recessive lethality of l(3)s2784 was due to effects on Doa expression, we mobilized the P-element insertion by screening for white− progeny in the F1 and recessive lethality to Df(3R)3450 in the F2. We sequenced 2 of 15 homozygous viable revertants recovered. Both showed precise excision of the l(3)s2784 P element and were viable in combination with two Doa alleles (HD and γ3B), demonstrating that the recessive lethality of l(3)s2784 is due to mutation of Doa. We conclude that the 105-kDa DOA protein is vital.
GAL4-directed expression reveals independent functions of the 55- and 69-kDa vs. the 105-kDa DOA isoforms:
We overexpressed specific Doa transcripts using the GAL4-UAS system (Brand and Perrimon 1993). EP and EY P elements are inserted, respectively, at the transcription start site of the entire 55-kDa coding transcript, just following noncoding exon N1 of the 55-kDa coding transcript, and at the 5′ end of the 105-kDa coding transcript (Figure 1, Table 1). None of these lines have any phenotypes on their own. We also constructed transgenic lines expressing the 69-kDa protein from cDNAs with long or truncated 3′-UTRs (Kpebe and Rabinow 2008). Isoform specificity for the P-element insertions permitting GAL4-directed Doa expression was confirmed on Northern blots (Figure 2B).
When expressed using the ubiquitous driver da-GAL4, no phenotypes are observed with any of the insertion alleles, aside from slight eye roughness and ectopic wing venation with P{EP}DoaEP3080 (not shown; Table 2). In contrast, the UAS-Doa69kD constructs with both long and short 3′-UTRs are lethal when expressed under the control of da-GAL4 (Table 2) (Kpebe and Rabinow 2008). Therefore, no further attempts were made to rescue the lethality of Doa null alleles via ubiquitous expression of the UAS-Doa69kD constructs.
TABLE 2.
Summary of isoform-specific GAL4 UAS-directed Doa expression phenotypes for P-element insertions, a transgene, and an RNAi construct
| GAL4 driver | DoaUAS allele | Phenotype |
|---|---|---|
| da-GAL 4 | EP3080 | Occasional minor ommatidial disorganization; 17% of EP3080/GMR-GAL4 females show ectopic vein formation in anterior compartment of wing blade, between L1 and L2. |
| UAS-Doa69kD | Lethal (Kpebe and Rabinow 2008). | |
| EY11301 | No abnormal phenotypes. | |
| EY08857 | No abnormal phenotypes. | |
| UAS-105kD-IR | Lethal (two lines). | |
| UAS-138kD-IR | Lethal at 29°, no phenotype at 25°. | |
| GMR-GAL 4 | EP3080 | Fused facets, disorganized interommatidial lattice and photoreceptors, holes in lens. |
| UAS-Doa69kD | Similar to or stronger than EP3080, depending on line (Kpebe and Rabinow 2008). | |
| EY11301 | Identical to EP3080. | |
| EY08857 | Barely perceptible eye roughness, essentially wild type. | |
| UAS-105kD-IR | Rough, shiny eyes (two lines). | |
| UAS-138kD-IR | Similar to UAS-105kD-IR at 29°; females show disrupted interommatidial bristles at the posterior eye margin at 25°. | |
| Lsp2-GAL 4 | EP3080 | Male lethal when Lsp-GAL 4 females are crossed with EP3080 males. In the reciprocal cross, 18% male F1. |
| UAS-Doa69kD | Three of four long 3′-UTR constructs are male lethal with reduced viability of females; short 3′-UTR constructs show only slight reduction in overall viability (40% of total F1 instead of 50%), slightly greater effects on male viability (15–20% total F1 instead of 50%). | |
| EY11301 | Male lethal (both directions in reciprocal crosses). | |
| EY08857 | No abnormal phenotypes (same proportion males and females). | |
| UAS-105kD-IR | No abnormal phenotypes (two lines). | |
| UAS-138kD-IR | No abnormal phenotypes, 25° or 29°. |
All crosses using the GAL4-UAS system were performed at 29° to enhance phenotypes except as noted. Lines expressing the UAS-Doa69kD construct presented similar phenotypes, whether carrying full-length or truncated 3′-UTRs, but those with the full-length UTR were generally stronger in phenotype. Four lines were analyzed for the long 3′-UTR construct and 18 for the short 3′-UTR construct. Two lines expressing the RNAi construct for the 105-kDa coding transcript gave identical results. The expression of the UAS-105kD-IR lines under control of the da-GAL4 driver was also tested at 25°, with the same result (lethality). A single line expressing an interfering RNA for the 138-kDa isoform transcript was tested.
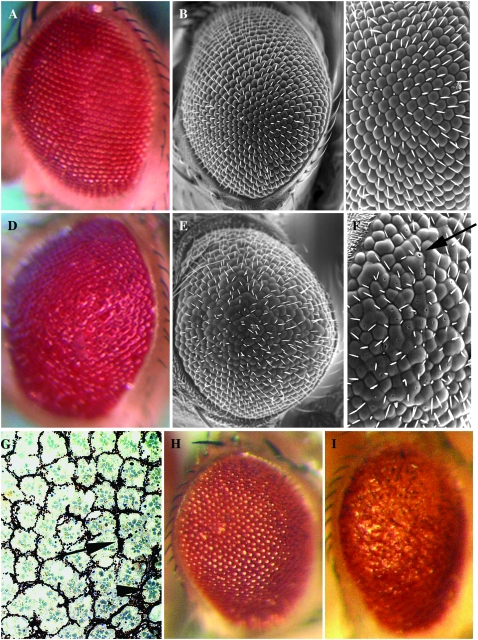
When driven with the GMR-GAL4 element, P{EP}DoaEP3080 and P{EPgy2}DoaEY11301 induce identical phenotypes, in which substantial ommatidial fusion and disorganization are observed (Figure 3, D–G; compare with siblings carrying a balancer chromosome and the GMR-GAL4 construct, Figure 3, A–C). Small holes in the surface of the ommatidia are reproducibly observed at high magnification in both these lines, which overexpress the 55-kDa kinase (arrow in Figure 3F), suggesting that cone cells either failed to fuse or failed to secrete sufficient lens proteins in these genotypes. Similar eye phenotypes are also observed with the lines expressing the UAS-Doa69kD construct, but the severity of the effects is dependent upon the specific line used (Kpebe and Rabinow 2008). Sagittal sections of a GMR-GAL4/+ ; DoaEP3080 fly (Figure 3G) show the fused ommatidia observed superficially (Figure 3, A–C), but also reveal supernumerary pigment cells (Figure 3G, arrow), and rhabdomeres (Figure 3G, arrowhead), suggesting that too many photoreceptor cells are present.
Figure 3.—
Eye phenotypes induced by GMR-GAL4-directed expression of specific DOA isoforms. (A–C) Optical and scanning electron microscopy of wild-type eyes shows the highly organized arrangement of the ommatidia and interommatidial bristles at both low (A and B) and high (C) magnification. These flies are genotype GMR-GAL4/+; TM6/+ siblings of the flies in D–F, which are genotype GMR-GAL4/+; DoaEP3080/+ and overexpress the 55-kDa nuclear kinase isoform. Note the shiny surface (D), the fused ommatidia (E), and pinpoint holes at the apex of several ommatidia (e.g., arrow in F). (G) A tangential section of a GMR-GAL4/+; DoaEP3080/+ eye is shown. Excess pigment lattice cells (arrow) and photoreceptor rhabdomeres (arrowhead) are observed, suggestive of too many cells or lack of cell death. Other ommatidia are fused and lack intervening pigment lattice cells. Identical disorganization of the eye is also observed in GMR-GAL4/+; DoaEY11301/+ individuals (not shown). These flies also overexpress the 55-kDa kinase, but lack its normal 5′-UTR (Figures 1 and 2). In contrast to these two lines, GMR-GAL4/+; DoaEY08857/+ eyes, expressing the 105-kDa cytoplasmic kinase, show no ommatidial disorganization (not shown). (H) GMR-GAL4/+; TM3/+, a sibling of (I) GMR-GAL4/+; DoaIR31049-MIMI/+, expressing an RNAi form of the 105-kDa coding transcript. The fused facets and general ommatidial disorganization resemble those of overexpression of the 55-kDa kinase. A second line carrying the same construction, DoaIR31049-FIMI, shows a virtually identical phenotype (not shown).
In contrast to these results, the eyes of flies expressing the 105-kDa cytoplasmic kinase via P{EPgy2}DoaEY08857 and GMR-GAL4 are essentially wild type (not shown). This observation supports the hypothesis that the cytoplasmic 105-kDa kinase isoform has a function distinct from the 55- and 69-kDa kinases.
Male-specific lethality induced by overexpression of the 55-kDa isoform in the larval fat body:
Expression of the 105-kDa kinase under control of the Lsp2-GAL4 driver also produced no evident phenotypes. However, both P{EP}DoaEP3080 and P{EPgy2}DoaEY11301 produce male-specific lethality when expressed with this driver (Table 2, supplemental Table 3). Male lethality is complete in the case of P{EPgy2}DoaEY11301, regardless of the direction of the cross, whereas for P{EP}DoaEP3080, male lethality is virtually complete when the Lsp2-GAL 4 driver is introduced from females. In the reciprocal cross, 18% of the progeny (instead of the 50% expected) are male. No abnormal phenotypes are observed in escaping males. The lethal period occurs during early development, since no dead larvae or pupae were observed on the vial walls. We also tested the UAS-69kD cDNA constructs with the Lsp2-GAL4 driver and observed reduced male survival, but female survival was also affected, albeit to a lesser degree. We therefore cannot be certain that expression of the 69-kDa kinase via the Lsp2-GAL4 element induces male-specific lethality, as is the case for EP(3)3080 and EY11301.
To investigate possible causes of male-specific lethality, we analyzed expression of the yolk protein genes (YP1, -2, and 3) and Sex-lethal (Sxl) by RT–PCR in male survivors of the Lsp2-GAL 4 × female P{EP}DoaEP3080 cross. YP genes, normally expressed only in female fat bodies, were analyzed to determine if the female sex-determination pathway was being activated by expression of the Doa 55-kDa isoform. Ectopic activation of Sxl and subsequent effects on male dosage compensation would be expected to result in male-specific lethality. However, inappropriate expression of the surveyed genes was not observed (not shown).
We also asked whether male-specific lethality due to overexpression of the DOA 55-kDa isoform from DoaEP3080 via the Lsp2-GAL4 driver was suppressed via the coexpression of TRA protein from a UAS-traF construct (Ferveur et al. 1995). We anticipated a negative result, since males often display increased sensitivity to detrimental mutations, in comparison with their female siblings, revealed via their reduced viability. However, somewhat surprisingly, TRA coexpression rescued male lethality, and equal numbers of both males and females eclosed. Thus, the activation of one or more female-specific genes in the fat body by the somatic sex-determination cascade is sufficient to suppress male-specific lethality induced by overexpression of the DOA 55-kDa isoform. Inappropriate levels of DOA kinase therefore appear to induce lethality due to effects on male-specific physiology and not due to interference with dosage compensation. Another possible explanation is that excess levels of TRA, a DOA substrate, titrate excess kinase and block its deleterious effects. In either case, the difference in male survival when comparing overexpression of the 55- and 105-kDa isoforms provides further support for the hypothesis that these proteins perform different functions.
Interfering RNA lines specific for the 105- and 138-kDa coding transcripts:
Due to a generous gift from Ryu Ueda and colleagues, we obtained two lines (UAS-105kD-IR) expressing an interfering RNA specific for 4.3-kb transcript encoding the 105-kDa cytoplasmic DOA isoform (Kpebe and Rabinow 2008). Consistent with the recessive lethality of l(3)s2784, which specifically reduces levels of this 4.3-kb transcript, expression of these constructs under control of the da-GAL4 ubiquitous driver results in complete and early lethality of all progeny, even at 25° (Table 2). GMR-GAL4-directed expression of both UAS-105kD-IR lines produces rough, shiny eyes with fused facets, reminiscent of the overexpression of the 55-kDa isoform in both EP3080 and EY11301 (Figure 3, H and I, compare Figure 3D with 3I). In contrast, expression of these RNAi constructs using the Lsp2-GAL4 produced no phenotype.
A single line expressing an interfering RNA for the 138-kDa DOA isoform (CG33204), obtained from the VDRC, was also tested with the GAL4 lines (Table 2). No phenotypes were observed at 25° with da-GAL4, whereas at 29° this combination was lethal in both sexes prior to the third instar. The UAS-138kD-IR construct produced no phenotypes at either 25° or 29°, aside from an occasional ectopic wing vein with the Lsp2-GAL4 driver. Expression of this construct with the GMR-GAL4 driver at 25° resulted in slight disruption of interommatidial bristles at the posterior edge of the eye, exclusively in females. At 29° with GMR-GAL4, a phenotype comparable to that of the UAS-105kD-IR lines was obtained with the UAS-138kD-IR construct, but exclusively in females (not shown). Interestingly, males were virtually unaffected. These results further support the hypothesis of differentiable functions for the 105-kDa protein vs. the 55- and 69-kDa proteins. The 138-kDa protein also seems to have a different role from the 55- and 69-kDa proteins, but may or may not be different from the 105-kDa protein.
No apparent roles for the DOA 105- and 138-kDa isoforms in somatic sex determination:
Expression of UAS-TRA-F under control of the escargot-GAL4 element induces strong male-to-female transformation and a dsx phenocopy in XY males, whereas expression of a UAS-TRA2-IR construct using this driver in XX females induces a nearly complete female-to-male sexual transformation typical of tra2 homozygous mutants (not shown). Since heteroallelic combinations of several Doa alleles induce female-to-male sex transformations, blocking production of the 105- or 138-kDa kinases would be thought to produce female-to-male sex transformations if either isoform has an important function in sex determination. However, expression of either the UAS-105kD-IR or the UAS-138kD-IR lines using the esg-GAL4 driver did not result in any detectable sex transformations. A “wings-down” phenotype and some ectopic wing venation were observed in the former line (not shown). Given the strong phenotypes produced in both sexes by these constructs when ubiquitously expressed or expressed in the eye, their failure to produce somatic sex transformations argues that neither the 105- nor the 138-kDa isoform plays an important role in somatic sex determination.
Complex complementation patterns among Doa alleles demonstrate pleiotropic phenotypes:
Inter se and reciprocal complementation tests were performed among 26 mutant Doa alleles, the majority of those available, to provide further insight into the functions of the locus. To summarize the results (Table 3, supplemental Table 4), our observations support the hypothesis that at least the 55- and 105-kDa isoforms define independent functions, since partially complementing alleles specifically affecting these isoforms produce phenotypes affecting different structures. Consistent with and extending previous observations, surviving heteroallelic adults possess missing or forked scutellar and head bristles, rough eyes, ectopic or incomplete wing veins, suppression of whiteapricot to wild-type levels, and subtle sex transformations. Dramatic female-to-male somatic sex transformation in two allelic combinations confirms previous observations on Doa's role in the somatic sex-determination cascade (Du et al. 1998) (Figure 4). Multiple allelic combinations with phenotypes specifically involving the tarsi were also observed, affecting only the metathoracic leg, with two exceptions (Figure 5). Strong maternal effects on the survival of heteroallelic combinations were noted in reciprocal crosses, suggesting the necessity for maternal contribution of the 105-kDa cytoplasmic protein to the oocyte (Table 4).
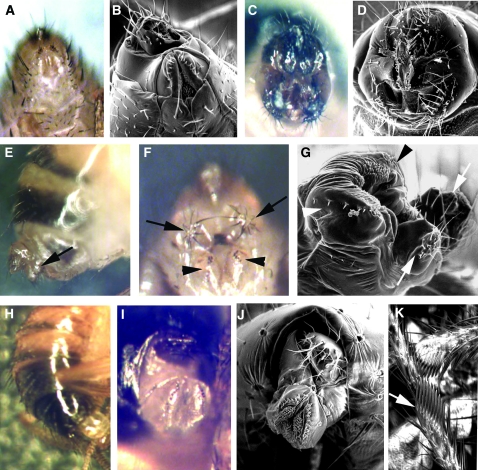
Figure 4.—
Partial complementation among specific Doa alleles leads to strong female-to-male somatic sex transformations. (A and B) Wild-type female genitalia, light and scanning electron microscopic views. (C and D) Wild-type male genitalia. (E–G) An XX, DoaE786/DoaEMS2 female: (E) lateral view, note the male “clasper” (arrow) but typical female pigmentation; (F and G) ventral view, symmetrical male claspers (arrows), as well as the female genital sex comb (arrowheads), produce a doublesex-like phenotype. (H–K) DoaEMS3/DoaHD female: (H) lateral view, note the male pigmentation; (I and J) ventral view, genitals are rotated but nontransformed; (K) perfectly formed male sex comb on a prothoracic leg (arrow).
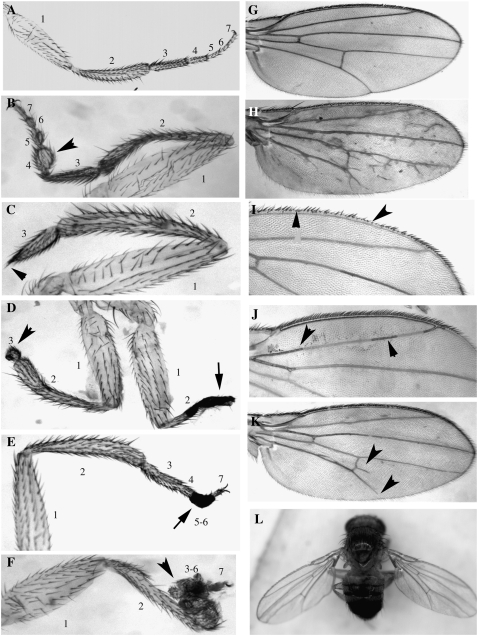
Figure 5.—
Examples of leg and wing phenotypes induced by heteroallelism for Doa alleles. (A) Wild-type metathoracic leg. 1, femur; 2, tibia; 3, tarsus 1; 4, tarsus 2; 5, tarsus 3; 6, tarsus 4; 7, tarsus 5. Segments distal to the first tarsus often fail to elongate (as in B, arrowhead), are missing (arrowheads in C and D), are necrotic (arrows in D and E), or are completely malformed (arrowhead in F). (B) DEM/l(3)01705; (C and D) EMS-C/EMS1; (E) EMS-C/γ3E; (F) EMS-C/E777. (G) Wild-type wing. Wings from Doa mutant combinations show sporadic ectopic veins (H), missing bristles along the anterior margin (I), blackened veins (J, arrowheads), or failure of L5 to attain the posterior wing margin (K, arrow). Note also the spur on the posterior cross-vein (arrowhead). Other allelic combinations display a “held-out” phenotype (L). (H) Su(Mrt9)/l(3)084402; (I) RemγA/E777; (J) EMS-2/γ3A; (K) RemγA/DEM; (L) 105/I5.
TABLE 4.
Nonreciprocal results observed in complementation tests
| Doa alleles | Female | Survival | Phenotype of heteroallelic survivors |
|---|---|---|---|
| 105/EMS-1 | 105 | + | Normal |
| EMS-1 | − | No survivors | |
| 105/EMS-3 | 105 | + | Only males survive, held-out wings, ectopic crossveins, missing scutellar bristles |
| EMS-3 | − | No survivors | |
| 105/I5 | 105 | + | Predominantly males observed, with rough eyes, held-out and fragile wings, postalar bristles duplicated, missing scutellar or ocellar bristles, slight sexual transformations of females: disorganization of vaginal bristles |
| I5 | − | No survivors | |
| 105/E777 | 105 | + | Only males observed, held-out wings, postalar bristles duplicated |
| E777 | − | No survivors | |
| 105/04743 | 105 | + | Only males observed, extremely rough eyes, ectopic wing veins, missing head, scutellar and thoracic bristles |
| 04743 | − | No survivors | |
| DEM/198-14 | DEM | − | No survivors |
| 198-14 | + | Postalar bristles duplicated, scutellar bristles forked-like | |
| DEM/04743 | DEM | + | Only rare pupae eclose, weak adults die quickly, rough eyes, missing bristles, incomplete L5 |
| 04743 | − | No survivors | |
| HD/s2784 | HD | + | Blackened areas on wings |
| S2784 | − | No survivors | |
| HD/EMS-3 | HD | + | Strong transformation of females toward males: male coloration of tergites 5 and 6, male sex combs on prothoracic leg, normal but rotated female genital plate |
| EMS-3 | − | No survivors | |
| EMS-1/Su(Mrt9) | EMS-1 | + | Small, rough eyes |
| Su(Mrt9) | − | No survivors | |
| EMS-1/γ3E | EMS-1 | + | Only males recovered, darkened areas in wing veins |
| γ3E | − | No survivors | |
| EMS-2/γ3A | EMS-2 | + | Badly oriented bristles on the head, missing or duplicated dorso-central bristles, forked scutellar bristles, fragile wings with incomplete posterior crossveins and darkened areas within veins |
| γ3A | − | No survivors | |
| CC/E786 | CC | + | Only females recovered, rough eyes, postalar bristles duplicated, missing scutellar bristles, disorganization of vaginal bristles |
| E786 | − | No survivors | |
| RemγA/γ3A | RemγA | + | Rough eyes, dorso-central or scutellar bristles missing, wings slightly curly, less translucent with small ectopic veins generally posterior to L2, disorganization of vaginal bristles |
| γ3A | − | No survivors | |
| RemγA/γ3C | RemγA | + | Rough eyes, fragile wings with little black spots (necrosis?), disorganization of vaginal bristles |
| γ3C | − | No survivors | |
| Msu2/γ3C | Msu2 | + | No phenotype observed |
| γ3C | − | No survivors | |
| Su(Mrt9)/Msu1 | Su(Mrt9) | + | Very rough eyes, some misoriented head bristles, disorganization of vaginal bristles |
| Msu1 | − | No survivors | |
| Su(Mrt9)/01705 | Su(Mrt9) | + | More females observed, rough eyes, head, scutellar or postalar bristles missing, fragile wings with small black spots and or ectopic veins, disorganization of vaginal bristles |
| 01705 | − | No survivors |
All heteroallelic survivors possessed wild-type red eyes due to suppression of wa to near normal pigment levels.
All Doa alleles suppress whiteapricot:
We had thought that Doa alleles obtained from mutagenic screens other than for suppression of wa might not possess this phenotype, since they might affect different functions. To our surprise this was not the case. All 24 Doa alleles tested dominantly suppress whiteapricot. Furthermore, complementing heteroallelic pairs suppress wa to full wild-type brick red (not shown), albeit with mosaicism in some combinations, probably due to particular alleles being associated with position effects.
Alleles producing peptides lacking the kinase catalytic domain are not complete nulls:
The RemγA allele is associated with chromosomal rearrangements and is due to position effect and not to a direct breakpoint within Doa (C. So and L. Rabinow, unpublished results), suggesting that it might generally reduce activity of all Doa isoforms. It behaves as a weak hypomorph, on the basis of its complementation properties (supplemental Table 3). To our surprise, this allele partially complements the kinase-null alleles γ3A and γ3C described above (supplemental Table 3). In contrast, a third kinase-null allele, γ3B, is not complemented. As described above, the former two alleles are due to small deletions introducing frameshifts and premature stop codons prior to the kinase catalytic domain, and they produce truncated peptides (Kpebe and Rabinow 2008). In contrast, the γ3B allele is due to a chromosomal breakpoint mapping upstream of the common domain (Yun et al. 1994), and no truncated peptides are recognized in γ3B/+ protein extracts (not shown). The partial complementation of both γ3A and γ3C by RemγA, but its failure to complement γ3B, suggests that the truncated peptides lacking the kinase catalytic domain present in the former two alleles are partially functional. This interpretation is supported by partial complementation of the EMS2 allele with γ3A, but not γ3B or γ3C (supplemental Table 3).
Sexual transformations in Doa heteroallelic combinations:
Dramatic female-to-male somatic sex transformations were observed in the allelic combination EMS2/E786, yielding a dsx-like phenotype in the genitalia, with both symmetrical male claspers and vaginal structures (Figure 4, E–G). This allelic combination does not produce male-like pigmentation on the female abdomen (Figure 4E) or male sex combs (not shown). The combination EMS3/HD also induces female-to-male somatic sex transformations but with different phenotypes, including male pigmentation on the fifth and sixth tergites (Figure 4H), rotated female genitalia (Figure 4, I and J), and male sex combs on the prothoracic legs (Figure 4K). In contrast with EMS2/E786, however, no male genital structures are observed. Other allelic combinations producing female-to-male transformations were less dramatic, in most cases demonstrating slight asymmetry in the female genital sex combs (Table 3, supplemental Table 4), as in DEM/HD heteroallelic mutants (Du et al. 1998). Ovaries dissected from some of the most dramatically transformed heteroallelic females (E786/DEM) were normal in appearance.
Doa affects formation of the mesothoracic tarsi:
The EMS-C allele is unique in that it is homozygous viable, fertile, and wild type in all phenotypes, aside from complete suppression of wa. Unsurprisingly, it complements all other Doa alleles, in combination with which it also suppresses wa to wild type. Minor phenotypes affect the bristles and wings in some of these combinations (supplemental Table 3). Effects of EMS-C in heteroallelic combinations are seen on the tarsi of the metathoracic legs, which are frequently deformed or missing (Figure 5). Similar deformities are occasionally observed in other Doa allelic combinations, e.g., 105/E786 and DEM/ l(3)01705. In almost all phenotypes involving leg development, the femur develops normally, and only weak deformities, if any, are observed in the tibia (e.g., Figure 5B). In contrast, metathoracic tarsal segments fail to elongate (Figure 5B), are necrotic or missing (Figure 5, D and E), or are fused and malformed (Figure 5F). Interestingly, the claws form normally in all cases. A single allelic combination, 105/E786, produces similar phenotypes in the mesothoracic tarsi, and the combination of l(3)01705/DEM affects both mesothoracic and metathoracic tarsi (supplemental Table 3). The cause of the difference in specificity of the alleles affecting the mesothoracic vs. the metathoracic tarsi is unclear, but these phenotypes demonstrate a requirement for Doa in morphogenesis of the distal leg segments.
Effects of Doa mutations on wing development:
Effects on wing structures and development were also noted. In Figure 5H, an example of a wing from a heteroallelic Doa fly displays numerous ectopic, branched, and divided veins, compared with wild type (Figure 5G). Different heteroallelic combinations induce missing bristles along the anterior wing margin (Figure 5I), blackened wing veins (Figure 5J), and spurs on the posterior cross-veins (Figure 5K). L5 systematically fails to reach the posterior wing margin in other combinations. The latter phenotype is observed in several allelic combinations involving the DEM allele. A “held-out” phenotype is observed in several genotypes involving the 105 allele (Figure 5L), suggesting a defect in thoracic musculature. These diverse phenotypes suggest roles for Doa in bristle, vein, and muscle formation.
Nonreciprocal complementation patterns among Doa alleles demonstrate maternal effects:
Nonreciprocal complementation was found among several alleles (Table 4), illustrated by their permissive role in the survival of heteroallelic adults. Many of these alleles are due to chromosomal rearrangements. Additionally, maternal effects of two P-element insertions near the start site of the 4.3-kb transcript suggest a role in oogenesis for the 105-kDa cytoplasmic protein.
Five alleles due to chromosomal rearrangements (105, CC, Msu1, Msu2, and RemγA) permit survival of a number of classes of heteroallelic adults if introduced from females but not from males (Table 4). The 105 allele in particular permits the viability of five alleles when introduced from females, but not in reciprocal crosses. Nonreciprocal complementation patterns with the rearrangement alleles even include the kinase-null alleles γ3A and γ3C.
In another example of a strong maternal effect on the survival of heteroallelic adults, the HD and l(3)s2784 alleles also complement each other in nonreciprocal patterns (Table 4). When l(3)s2784/TM6B males are crossed with HD/TM6B females, heteroallelic F1 survive and display no mutant phenotypes. In the reciprocal cross, heteroallelic mutants die at or prior to the pupal stage. Interestingly, the nearby P-element insertions l(3)04703, in combination with either the 105 or the DEM alleles, and l(3)01705, in combination with Su(Mrt9), also complement when introduced from males, while failing to support complementation when introduced from the females. Taken together, these observations suggest that the P elements located at or near the initiation site of the 4.3-kb transcript coding the 105-kDa cytoplasmic DOA kinase interfere with a process required for oogenesis or early larval development. In addition, the chromosomal rearrangement alleles cited would produce levels of this transcript sufficient to allow maternal rescue and survival through adulthood.
DISCUSSION
Constraints on the structure of the DOA kinase catalytic and noncatalytic domains:
Despite our inability to localize any point or small sequence changes causing isoform-specific Doa mutations, SSCP analysis of multiple alleles from various genetic backgrounds illustrates the extreme amino acid sequence conservation imposed on the LAMMER kinase common domain. This hypothesis is reinforced by high levels of amino acid identity observed in kinase catalytic domain coding sequences in LAMMER kinase orthologs from distantly related species, e.g., 96% between D. melanogaster and D. virilis (Kpebe and Rabinow 2008) and 75% between D. melanogaster and CLK2 of Homo sapiens (Yun et al. 1994).
These observations contrast with the relative relaxation of sequence constraints observed in the N-terminal noncatalytic domains, as evidenced by the multiple-amino-acid substitutions, insertions, and deletions found among the laboratory strains surveyed. The degree of polymorphism in the N-terminal domains in these laboratory strains is in fact somewhat surprising given the 51–85% amino acid sequence identity we found in comparing N-terminal domain sequences of D. melanogaster with the distantly related D. virilis (Kpebe and Rabinow 2008). It seems that natural selection, reduced among laboratory populations, must keep these variations in check in the wild.
Our inability to identify lesions provoking many Doa alleles suggests that they lie either in the extensive noncoding sequences of the gene, affecting its transcription or splicing, or in exons coding the N termini of the 227- and the 138-kDa isoforms, which were not surveyed. Of the two possibilities, the latter seems less likely given the overall size of the gene, but we cannot exclude any alternatives. We also cannot rule out the possibility that transcripts and proteins exist beyond those that we identified, despite our extensive characterization of the gene's products (Kpebe and Rabinow 2008).
Partial complementation by kinase-null alleles implies functions for the noncatalytic N termini:
We identified the lesions causing kinase-null mutations that introduce premature termination codons within the common domain and accumulate detectable levels of truncated peptides in the γ3A and γ3C alleles (Kpebe and Rabinow 2008), in contrast to the γ3B allele. Complementation tests with these three alleles reveal that γ3A and γ3C both partially complement some hypomorphic alleles. This partial complementation suggests that the N-terminal noncatalytic peptides possess partial function. These functions could be involved in dimerization with the full-length kinase, its regulation or localization, or other protein–protein interactions.
A phenotype common to all Doa alleles:
Most Doa mutations were isolated as suppressors of whiteapricot. We anticipated that some of the alleles isolated in other screens would not have this phenotype, since they might affect other isoforms of the kinase with different functions. However, six alleles obtained via other types of screens all suppress wa. Thus, all the isoforms mutated in our collection possess at least one phenotype in common.
Somatic sex transformations induced by different combinations of Doa alleles produce different phenotypes:
Multiple combinations of Doa alleles produce very slight female-to-male somatic sex transformations, principally resulting in minor disruption of the vaginal sex comb, as previously reported for the HD/DEM combination (Du et al. 1998). We observed strong sex transformations in two new Doa heteroallelic genotypes. In the first case a dsx-like phenotype was observed, with male and female genital structures present, although without male pigmentation on the fifth and sixth tergites or formation of male sex combs on the prothoracic legs. In a second instance, genital plate rotation, male abdominal pigmentation, and formation of male sex combs were observed, while no genital transformation occurred. Thus, even when partially complementing alleles affect the same process, different allelic pairs produce differentiable phenotypes. These observations suggest either that these alleles affect specific domains or isoforms of the kinase interacting with different partners or that these mutations affect expression of the kinase at different times and places.
Doa isoforms with separable functions:
A contrast to the suppression of wa demonstrated by all alleles is offered by the complementation patterns of alleles specifically affecting the 2.7-kb transcripts generating 55- and 69-kDa proteins, vs. the P-element insertions affecting the 4.3-kb transcript, coding the 105-kDa protein. The 55-kDa protein is at least partially nuclearly localized, whereas the 105-kDa protein is exclusively cytoplasmic during pupation (Yun et al. 2000). Our data strongly suggest that these two isoforms serve different functions.
The copia insertion in the HD allele primarily reduces levels of the 2.7-kb transcripts coding the 55- and 69-kDa proteins, and this allele is involved in several combinations producing sex transformations. GAL4-driven expression of the 55-kDa kinase also produces phenotypes including eye roughness and male-specific lethality. The intracellular localization of the 55-kDa protein matches the presumed nuclear localization of TRA and TRA2, although the distribution of the latter proteins has only been demonstrated through gene fusions. Thus, we suggest that sex-determination defects in Doa mutants, due to hypophosphorylation of SR and SR-like proteins such as TRA and TRA2, are due to reduced levels of the 2.7-kb transcripts and the 55- and 69-kDa kinases.
In contrast, RNAi constructs and alleles reducing levels of the 4.3-kb transcript and hence the 105-kDa kinase do not seem to produce sex transformations. Expression of an interfering RNA for this isoform in the genitals failed to provoke even slight sex transformations, although the same constructs produced phenotypes when expressed in the whole fly and in the eye. Additionally, no phenotypes were detected due to overexpression of the 105-kDa kinase, unlike the phenotypes observed for the 55- and 69-kDa kinases. Nevertheless, the 105-kDa protein is vital to the fly, since the recessive lethality of at least l(3)s2784 is due to the insertion in Doa, as proven by the recovery of homozygous viable revertants when the P-element insertion was excised. Moreover, interfering RNAs targeting this isoform are lethal if widely expressed.
Phenotypes induced by GAL4-driven expression of an interfering RNA for the 138-kDa protein are similar to those observed for the 105-kDa protein, including lethality when widely expressed, rough eyes (albeit female specific), and no apparent effects on sex determination.
In sum, we conclude that at a bare minimum the 55- and 105-kDa kinases serve clearly different functions. Further experiments will be needed to determine whether the 69- and 138-kDa kinases significantly overlap the former two kinases in their functions.
Maternal effects of Doa alleles suggest a specific role for the 105-kDa protein in oogenesis or early embryonic development:
Crosses producing nonreciprocal results by definition demonstrate maternal effects for Doa. To summarize, mutations affecting the 4.3-kb transcript are less likely to produce heteroallelic adults when introduced from the mother, vs. maternal contribution of the HD allele, which affects the 2.7-kb transcripts.
We interpret these results in light of the analyses conducted in previous studies (Yun et al. 2000; Kpebe and Rabinow 2008). The 4.3-kb mRNA is not expressed in embryos, although the 105-kDa protein it encodes is found on immunoblots in extracts from 0- to 4-hr-old embryos. In contrast, high levels of the 2.7-kb transcripts are expressed in early embryos and throughout embryogenesis. Therefore, the ubiquitous 55-kDa isoform can be either maternally contributed or produced in the zygote, since its transcription is activated to high levels during early embryogenesis. In contrast, the 105-kDa cytoplasmic protein must be contributed maternally.
This observation suggests that the 105-kDa protein is essential for oogenesis and embryonic development, since the mothers in these crosses are heterozygous and would be expected to produce normal levels of the kinase from their balancer chromosome. Support for the necessity of the 105-kDa protein in oogenesis is also derived from germ-line clonal analysis of loci on 3R with maternal effects required for zygotic development (Perrimon et al. 1996), in which the Doa allele l(3)01705 was examined. No germ-line clones were recovered, similar to our results using the γ3B (null) allele (Yun et al. 2000). The insertion site of the P element in l(3)01705 is only 5 bp distant from l(3)s2784, suggesting that that the former allele specifically affects the 4.3-kb transcript, as does the latter.
Male-specific lethality induced by overexpression of the 55-kDa kinase in the larval fat body:
GAL4-directed expression of the 55-kDa kinase in the larval fat body results in male-specific lethality for reasons we were unable to determine. Curiously, male-specific lethality induced by expression of the 55-kDa kinase in larval fat body is rescued by coexpression of TRA protein. While we cannot offer an explanation for these observations, we note that fat body physiology and gene expression are implicated in several aspects of sexual dimorphism (Dauwalder et al. 2002; Fujii and Amrein 2002; Lazareva et al. 2007). Further investigation will be necessary to identify the specific targets of the 55-kDa kinase necessary to produce male-specific lethality.
Acknowledgments
Drosophila lines were kindly provided by Jason Morris, Steve Mount, Brad Philipps, Jean-René Martin, Bill Mattox, Stephanie LeBras, and Eyal Schejter, as well as by the Drosophila Stock Centers at Indiana University and Szeged, Hungary, the Baylor University Drosophila Genome Project, and the Vienna Drosophila RNAi stock center. Special thanks are due to Jim Birchler for previously unpublished Doa alleles and to Ryu Ueda and colleagues of the Japanese National Institute of Genetics, Genetic Strains Research Center, for generously providing RNAi lines specific for the 105-kDa DOA isoform (CG31049) prior to public release. We also thank Severine Domenichini (Plateforme Microscopie Electronique, Imagerie-Biocellulaire IFR87, Orsay-Gif, Université Paris 11) for scanning electron micrography; and Danielle Jaillard and Jeril Degrouard, of the Centre National de la Recherche Scientifique (CNRS) Unité Mixte de Recherche C8080 electron microscopy facility, for help with retinal sections; and Bill Mattox and Brian Oliver for helpful discussions. Financial support was provided by the CNRS and the Université Paris Sud 11. Additional funding was provided by the French Ministry of Research and the CNRS (03G138), in the Biological Resource Center program for fundamental research, “Natural and induced variants in Drosophila,” and by the Division de la Recherche, Université Paris 11.
References
- Bard, F., L. Casano, A. Mallabiabarrena, E. Wallace, K. Saito et al., 2006. Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature 439 604–607. [DOI] [PubMed] [Google Scholar]
- Ben-David, Y., K. Letwin, L. Tannock, A. Bernstein and T. Pawson, 1991. A mammalian protein kinase with potential for serine/threonine and tyrosine phosphorylation is related to cell cycle regulators. EMBO J. 10 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias, M., R. Giet, R. Sinka, A. Mazumdar, W. G. Lock et al., 2004. Genome-wide survey of protein kinases required for cell cycle progression. Nature 432 980–987. [DOI] [PubMed] [Google Scholar]
- Bjorklund, M., M. Taipale, M. Varjosalo, J. Saharinen, J. Lahdenpera et al., 2006. Identification of pathways regulating cell size and cell-cycle progression by RNAi. Nature 439 1009–1013. [DOI] [PubMed] [Google Scholar]
- Brand, A., and N. Perrimon, 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118 401–415. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega, J. A., and V. Hartenstein, 1997. The Embryonic Development of Drosophila melanogaster. Springer, Berlin.
- Campuzano, S., L. Balcells, R. Villares, L. Carramolino, L. Garcia-Alonse et al., 1986. Excess function Hairy-wing mutations caused by gypsy and copia insertions within structural genes of the achaete-scute locus of Drosophila. Cell 44 303–312. [DOI] [PubMed] [Google Scholar]
- Colwill, K., T. Pawson, B. Andrews, J. Prasad, J. L. Manley et al., 1996. The Clk/STY protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 15 265–275. [PMC free article] [PubMed] [Google Scholar]
- Dauwalder, B., S. Tsujimoto, J. Moss and W. Mattox, 2002. The Drosophila takeout gene is regulated by the somatic sex-determination pathway and affects male courtship behavior. Genes Dev. 16 2879–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierick, H. A., and R. J. Greenspan, 2006. Molecular analysis of flies selected for aggressive behavior. Nat. Genet. 38 1023–1031. [DOI] [PubMed] [Google Scholar]
- Dietzl, G., D. Chen, F. Schnorrer, K. C. Su, Y. Barinova et al., 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448 151–156. [DOI] [PubMed] [Google Scholar]
- Du, C., M. E. McGuffin, B. Dauwalder, L. Rabinow and W. Mattox, 1998. Protein phosphorylation plays an essential role in the regulation of alternative splicing and sex determination in Drosophila. Mol. Cell 2 741–750. [DOI] [PubMed] [Google Scholar]
- Edwards, A. C., S. M. Rollmann, T. J. Morgan and T. F. Mackay, 2006. Quantitative genomics of aggressive behavior in Drosophila melanogaster. PLoS Genet. 2 e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferveur, J. F., K. F. Stortkuhl, R. F. Stocker and R. J. Greenspan, 1995. Genetic feminization of brain structures and changed sexual orientation in male Drosophila. Science 267 902–905. [DOI] [PubMed] [Google Scholar]
- Forch, P., and J. Valcarcel, 2003. Splicing regulation in Drosophila sex determination. Prog. Mol. Subcell. Biol. 31 127–151. [DOI] [PubMed] [Google Scholar]
- Fujii, S., and H. Amrein, 2002. Genes expressed in the Drosophila head reveal a role for fat cells in sex-specific physiology. EMBO J. 21 5353–5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski, S. M., S. Chittaranjan, E. D. Pleasance, J. D. Freeman, C. L. Anderson et al., 2003. A SAGE approach to discovery of genes involved in autophagic cell death. Curr. Biol. 13 358–363. [DOI] [PubMed] [Google Scholar]
- Howell, B. W., D. E. H. Afar, J. Lew, E. M. J. Douville, P. L. E. Iceley et al., 1991. STY, a tyrosine-phosphorylating enzyme with sequence homology to serine/threonine kinases. Mol. Cell. Biol. 11 568–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kpebe, A., and L. Rabinow, 2008. Alternative promoter usage generates multiple evolutionarily conserved isoforms of Drosophila DOA kinase. Genesis 46 132–143. [DOI] [PubMed] [Google Scholar]
- Lazareva, A. A., G. Roman, W. Mattox, P. E. Hardin and B. Dauwalder, 2007. A role for the adult fat body in Drosophila male courtship behaviour. PLoS Genet. 3 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C. Y., E. A. Clough, P. Yellon, T. M. Teslovich, D. A. Stephan et al., 2003. Genome-wide analyses of steroid- and radiation-triggered programmed cell death in Drosophila. Curr. Biol. 13 350–357. [DOI] [PubMed] [Google Scholar]
- Lee, K., C. Du, M. Horn and L. Rabinow, 1996. Activity and autophosphorylation of LAMMER protein kinases. J. Biol. Chem. 271 27299–27303. [DOI] [PubMed] [Google Scholar]
- Morris, J. Z., C. Navarro and R. Lehmann, 2003. Identification and analysis of mutations in bob, Doa and eight new genes required for oocyte specification and development in Drosophila melanogaster. Genetics 164 1435–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayler, O., S. Stamm and A. Ullrich, 1997. Characterization and comparison of four serine- and arginine-rich (SR) protein kinases. Biochem. J. 326 693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolakaki, E., C. Du, J. Lai, L. Cantley, T. Giannakouros et al., 2002. Phosphorylation by LAMMER protein kinases: determination of a consensus site, identification of in vitro substrates and implications for substrate preferences. Biochemistry 41 2055–2066. [DOI] [PubMed] [Google Scholar]
- Norga, K. K., M. C. Gurganus, C. L. Dilda, A. Yamamoto, R. F. Lyman et al., 2003. Quantitative analysis of bristle number in Drosophila mutants identifies genes involved in neural development. Curr. Biol. 13 1388–1396. [DOI] [PubMed] [Google Scholar]
- O'Connell, P. O., and M. Rosbash, 1984. Sequence, structure and codon preference of the Drosophila ribosomal protein 49 gene. Nucleic Acids Res. 12 5495–5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J. W., K. Parisky, A. M. Celotto, R. A. Reenan and B. R. Graveley, 2004. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc. Natl. Acad. Sci. USA 101 15974–15979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon, N., A. Lanjuin, C. Arnold and E. Noll, 1996. Zygotic lethal mutations with maternal effect phenotypes in Drosophila melanogaster. II. Loci on the second and third chromosomes identified by P-element-induced mutations. Genetics 144 1681–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad, J., K. Colwill, T. Pawson and J. L. Manley, 1999. The protein kinase Clk/Sty directly modulates SR protein activity: both hyper- and hypophosphorylation inhibit splicing. Mol. Cell. Biol. 19 6991–7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinow, L., and J. A. Birchler, 1989. A dosage-sensitive modifier of retrotransposon induced alleles of the Drosophila white locus. EMBO J. 8 879–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinow, L., S. L. Chiang and J. A. Birchler, 1993. Mutations at the Darkener of apricot locus modulate transcript levels of copia and copia-induced mutations in Drosophila melanogaster. Genetics 134 1175–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone, G., A. Pane and L. C. Polito, 2002. Sex determination in flies, fruitflies and butterflies. Genetica 116 15–23. [DOI] [PubMed] [Google Scholar]
- Savaldi-Goldstein, S., G. Sessa and R. Fluhr, 2000. The ethylene-inducible PK12 kinase mediates the phosphorylation of SR splicing factors. Plant J. 21 91–96. [DOI] [PubMed] [Google Scholar]
- Tautz, D., and C. Pfeifle, 1989. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98 81–85. [DOI] [PubMed] [Google Scholar]
- Villard, L., A. Kpebe, C. Cardoso, P. J. Chelly, P. M. Tardieu et al., 2000. Two affected boys in a Rett syndrome family: clinical and molecular findings. Neurology 55 1188–1193. [DOI] [PubMed] [Google Scholar]
- Yun, B., R. Farkas, K. Lee and L. Rabinow, 1994. The Doa locus encodes a member of a new protein kinase family, and is essential for eye and embryonic development in Drosophila melanogaster. Genes Dev. 8 1160–1173. [DOI] [PubMed] [Google Scholar]
- Yun, B., K. Lee, R. Farkas, C. Hitte and L. Rabinow, 2000. The LAMMER protein kinase encoded by the Doa locus of Drosophila is required in both somatic and germ-line cells, and is expressed as both nuclear and cytoplasmic isoforms throughout development. Genetics 156 749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachar, Z., D. Davison, D. Garza and P. M. Bingham, 1985. A detailed developmental and structural study of the transcriptional effects of insertion of the copia transposon into the white locus of Drosophila melanogaster. Genetics 111 495–515. [DOI] [PMC free article] [PubMed] [Google Scholar]