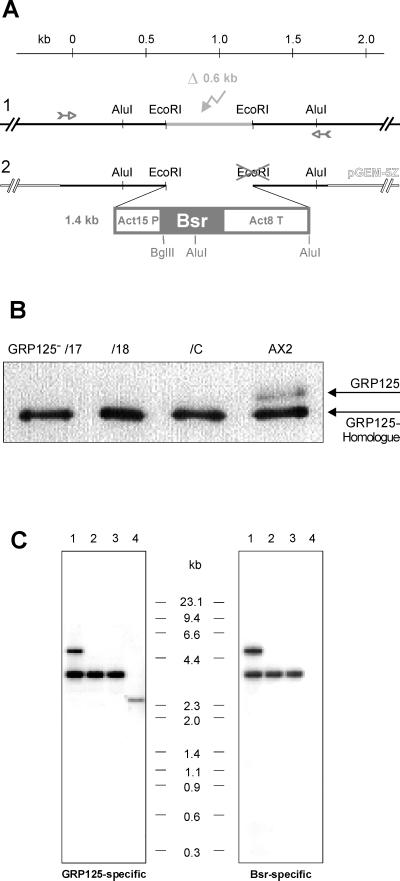
Figure 7.
(A) Schematic presentation of the replacement strategy yielding the inactivation of the GRP125 gene in Dictyostelium. The 1.8-kb genomic region at the 5′ end of the GRP125 gene was amplified via PCR (small arrows) and cloned into pGEM-5Z. The EcoRI–EcoRI-fragment (light gray) was deleted (flash in light gray) (1) and replaced by blunt-end insertion of the Bsr resistance cassette (2), Act15P, actin 15 promotor; Act8T, actin 8 terminator; Bsr, blasticidin-S deaminase gene. Restriction sites marked in gray got lost (cross) or introduced in the replacement construct. A kilobase ruler refers to nucleotide positions in the GRP125 gene (A of ATG = 1). (B) Western blot analysis of homogenates of three independent GRP125-deficient mutants (GRP125−/17, GRP125−/18, and GRP125−/C)and AX2 cells. Cell lysates (1 × 106 cells, growth phase) were analyzed with the polyclonal anti-GRP125/S3S4 antibody. GRP125−/17, GRP125−/18, and GRP125−/C fail to express GRP125 but produce the GRP125 homologue. (C) Comparative Southern blot analysis of genomic DNA of GRP125-deficient cells (GRP125−/17 [1], GRP125−/18 [2], and GRP125−/C [3]) and AX2 cells (4). Genomic DNA from D. discoideum AX2 and the mutants was EcoRI–BglII double digested, size fractionated by electrophoresis in agarose gels, and transferred to a Hybond-N membrane. The blot was hybridized to radioactively labeled DNA probes of GRP125 corresponding to a fragment exactly 3′ of the deleted EcoRI–EcoRI fragment (GRP125-specific probe F1, bp 1228–2727 of the GRP125 coding region) or the Bsr encoding sequence.