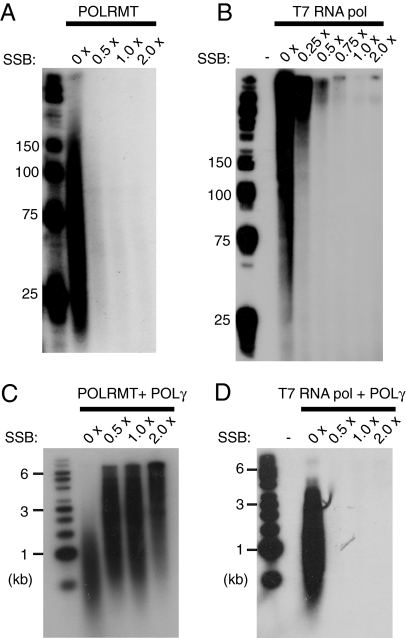
Fig. 4.
MtSSB influences POLRMT-dependent primer synthesis. (A) RNA synthesis on ssDNA was performed as described in Material and Methods. Reactions mixtures (25 μl) contained M13mp18 ssDNA (35 fmol), POLRMT (500 fmol), and increasing amounts of mtSSB (0 fmol, 8.4 pmol, 16.8 pmol, 33.6 pmol). Each mtSSB monomer covers approximately 60 nt (33, 34) and the saturation level for each protein concentration was calculated. The saturation level 1 × indicates that the ssDNA molecules should be completely covered with mtSSB. After the incubation for 1 h at 37°C, reaction products were separated by electrophoresis on a denaturing polyacrylamide gel (10%) and detected by autoradiography. (B) Reactions were performed as in (A), but in the presence of T7 RNA polymerase (1 unit). (C) RNA-primed DNA synthesis on ssDNA in the presence of [α-32P]dCTP for labeling of DNA. Reactions (25 μl) contained M13mp18 ssDNA (35 fmol), POLRMT (500 fmol), POLγA (100 fmol), POLγB (300 fmol), and increasing amounts of mtSSB [saturation levels are indicated in (A)]. The reactions were allowed to proceed for 1 h at 37°C and the reaction products were separated by electrophoresis on a denaturing agarose gel (1%) and detected by autoradiography. (D) Reactions were performed as in (C), but in the presence of T7 RNA polymerase (1 unit).