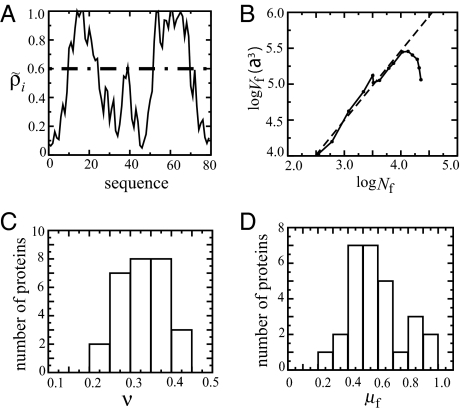
Fig. 2.
Scaling of the folded core with number of monomers. A and B correspond to λ−repressor (1lmb). (A) Residues with native density ρ̃i > 0.6 (indicated by the dashed line) define a fiducial set of folded residues. (B) Linear fit of log Vf vs. log Nf (dashed line) gives the exponent of Vf ≈ Nf3ν. In this example, the fitting equation is y = 5.6 + 0.97x, so that ν = 0.32, and b3 = 5.6a3; a = 3.8 Å is the average distance between the α carbons. (C) Histogram of scaling exponent ν for 28 proteins. (D) Histogram of the packing fraction of the folded core of the critical nucleus at Tf.