Abstract
Prokaryotic and eukaryotic replicons possess a distinctive region containing a higher than average number of adenine and thymine residues (the AT-rich region) where, during the process of replication initiation, the initial destabilization (opening) of the double helix takes place. In many prokaryotic origins, this region consists of repeated 13-mer motifs whose function has not yet been specified. Here we identify specific mutations within the 13-mer sequences of the broad-host-range plasmid RK2 that can result in defective origin opening or that do not affect opening but induce defects in helicase loading. We also show that after the initial recruitment of helicase at the DnaA-box sequences of the plasmid origin, the helicase is translocated to the AT-rich region in a reaction requiring specific sequence of the 13-mers and appropriate facing of the origin motifs. Our results demonstrate that specific sequences within the AT-rich region of a replication origin are required for either origin opening or helicase loading.
Keywords: 13-mer sequences, DNA replication, DnaB helicase, plasmid RK2
The initiation of DNA synthesis requires a mechanism to coordinate DNA unwinding, helicase recruitment, and site-specific helicase loading. Binding of the initiator (Rep), a single protein, or a complex of several proteins to the replication origin results in the melting of the double-stranded DNA helix in a region of the origin with low internal thermodynamic stability because of a higher than average content of adenine and thymine residues (AT-rich region). In plasmid replicons, the involvement of both the host-encoded replication initiator DnaA and the plasmid encoded Rep protein is essential for origin melting, helicase recruitment, and helicase loading onto the ssDNA in the opened AT-rich region.
DNA replication of the broad-host-range plasmid RK2 is initiated by the binding of the plasmid-encoded protein TrfA to direct repeats (iterons) present at its replication origin (oriV), a process similar to that of other plasmid systems (1, 2). The formation of an open complex at oriV by the TrfA initiation protein requires HU and is stabilized by the host DnaA protein (3). In contrast, the initiation of replication at the Escherichia coli chromosomal origin, oriC, involves a single initiator, DnaA, binding to DnaA-box sequences and resulting in duplex DNA melting (4–7). During replication initiation at RK2 oriV, helix destabilization occurs within the AT-rich region, which includes four 13-mer repeated sequences (L, M1, M2, R) that exhibit a DNA consensus sequence of TAAACnTTnTTTT (3). Similar motifs have been identified in a majority of theta-replicating plasmids and bacterial chromosomes including E. coli, where three 13-mers (L, M and R) have been identified in oriC (4–6). The position of the 13-mer clusters and their specific sequences are important for proper functioning of both oriV (8, 9) and oriC (5, 10, 11). Although the importance of 13-mers is well established, their exact role is not completely understood.
Certain E. coli proteins, including SeqA (12, 13), IciA (11), and DnaA (14, 15), specifically interact with the oriC AT-rich region. In addition, DnaB helicase and DnaC interact with ssDNA in the open complex (16, 17). Helicase loading at the opened oriC (18) requires the interaction of DnaA with DnaB (19). The nature of the interaction of DnaA with the AT-rich region is unclear. Earlier evidence indicated that DnaA, when bound with ATP, interacts with six-nucleotide sequences, termed ATP-DnaA boxes, within the AT-rich region (15, 20). More recent data suggest that the ATP-DnaA boxes are not the sites for DnaA interaction within the 13-mer region (21). When RK2 DNA replication is initiated in E. coli, DnaB helicase is recruited by DnaA to dsDNA containing oriV DnaA-box sequences located upstream of the 13-mer cluster (22). ATP-DnaA boxes are not present in oriV (3, 9). It has been shown that the TrfA protein is essential for the helicase complex activity (23); however, the mechanism of this activation has not yet been elucidated.
In this work, mutations in the 13-mers that result in defects in helicase loading, but not in origin opening, have been characterized and used for the analysis of helicase complex formation at the replication origin of the broad-host-range plasmid RK2.
Results and Discussion
DNA Unwinding Activity of E. coli DnaB Helicase Is Affected by Mutations in the AT-Rich Region.
In our previous study, 26 plasmid mutants with an altered sequence, position, or spacing of the 13-mer motifs of the RK2 replication origin were constructed and analyzed for replication activity (9). The data showed that both the sequence and the position of 13-mers are critical for oriV functionality. To determine what particular stage during the initiation of plasmid replication requires 13-mer sequence specificity, we analyzed in vitro the individual steps of replication initiation in these oriV mutants.
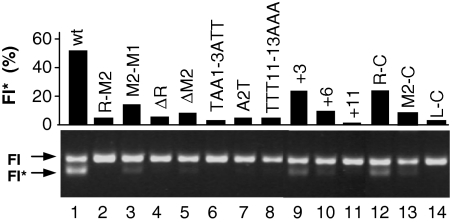
E. coli DnaB helicase complex activity on miniRK2 plasmid templates with either wild-type or altered 13-mer sequences was identified electrophoretically by the formation of a substantially unwound form of the supercoiled plasmid DNA, designated FI* (23). The formation of the FI* form is a result of the combined activities of plasmid and bacterial replication initiation proteins, including DnaA, DnaC, gyrase, SSB, HU, and TrfA. We optimized the reaction conditions so that the helicase activity was ≈50% on the wild-type template, as measured by densitometry scan of the gel (Fig. 1 lane 1). This allowed us to observe either an increase or a reduction of helicase activity of the oriV mutants. Thirteen mutants exhibiting no or substantially reduced helicase activity were selected (Fig. 1) (Table 1). No mutants with increased helicase activity in comparison to the wild-type template were identified. Because the TrfA protein exists in two forms: 33 kDa, allowing for replication in E. coli and 44 kDa, essential for replication in Pseudomonas, we performed FI* experiments by using either the short or long version of the protein. The effects were similar, regardless of the version of TrfA protein present in the reaction (data not shown); therefore, in subsequent experiments only the TrfA 33kDa form of the protein was used. The substitution of 13-mer M2 with M1 (mutant M2-M1) resulted in low helicase activity (Fig. 1, lane 3), whereas no activity was detected with mutant R-M2 (Fig. 1, lane 2). Deletion (Fig. 1, lanes 4 and 5) and insertion (Fig. 1 lanes 9–11) mutants exhibited only faint FI* activity. Helicase was also inactive on DNA templates where the sequence, but not AT-richness (the AT/GC ratio), was changed (Fig. 1, lanes 6–8). The substitution of the right (R) or the middle (M2) 13-mer with chromosomal E. coli oriC 13-mer (mutants R-C and M2-C, respectively) reduced the activity of DNA helicase, whereas no activity was detected if the left (L) 13-mer was substituted (Fig. 1, lanes 12–14).
Fig. 1.
DnaB helicase activity at oriV mutant templates. E. coli DnaB helicase activity on oriV mutant templates was determined by using the FI* assay. The position of the FI* DNA band, the extensively unwound covalently closed circular DNA generated by helicase activity on the supercoiled (FI) template DNA, is indicated by an arrow. The reactions were performed as described in Materials and Methods. DnaB at 600 ng and DnaC at 20 ng were used to optimized the reaction so that the helicase activity was ≈50% on the wild-type template. Black columns indicate the conversion to FI* form, calculated on the basis of densitometry analysis, as the percentage of total DNA in the reaction.
Table 1.
Effect of mutations within 13-mer motifs and oriV insertion mutants on origin replication activity
| oriV mutant | Sequence of mutated AT-rich region or insertion |
In vivo |
In vitro |
|||
|---|---|---|---|---|---|---|
| TF | FII | FI* | KMnO4 | EM | ||
| wt | TAACCTGTCTTTTAACCTGCTTTTAAACCAATATTTATAAACCTTGTTTT | + | + | + | + | + |
| R-C | TAACCTGTCTTTTAACCTGCTTTTAAACCAATATTTAGATATCTTATTAG | − | − | − | + | +/− |
| M2-C | TAACCTGTCTTTTAACCTGCTTTTGATATCTTATTAGTAAACCTTGTTTT | − | +/− | − | + | − |
| L-C | GATATCTTATTAGAACCTGCTTTTAAACCAATATTTATAAACCTTGTTTT | − | − | − | − | nt |
| R-M2 | TAACCTGTCTTTTAACCTGCTTTTAAACCAATATTTATAAATATTGGTTT | − | − | − | + | − |
| M2-M1 | TAACCTGTCTTTTAACCTGCTTTTAAAAAGCAGGTTATAAACCTTGTTTT | − | +/− | +/− | + | − |
| ΔR | TAACCTGTCTTTTAACCTGCTTTTAAACCAATATTTATAA. . . . . . . . . . | − | − | − | +/− | nt |
| ΔM2 | TAACCTGTCTTTTAACCTGCTTTTA. . . . . . . . . . . . TAAACCTTGTTTT | − | − | − | +/− | nt |
| A2T | TTACCTGTCTTTTTACCTGCTTTTAAACCAATATTAATTAACCTTGTTTT | − | − | − | − | nt |
| TAA1–3ATT | ATTCCTGTCTTTTTTCCTGCTTTTAAACCAATATTTAATATTCTTGTTTT | − | − | − | − | nt |
| TTT11–13AAA | TAACCTGTCTAATAACCTGCTTAAATTCCAATATTTATAAACCTTGTAAA | − | − | − | + | − |
| +3 | AGA | − | − | − | − | nt |
| +6 | GAATTC | − | − | − | − | nt |
| +11 | GTAGATACAAG | − | − | − | − | nt |
| oriV3–6 | GAATTC | − | − | − | − | − |
| oriV3–11 | GAATTCGTCAG | + | + | + | nt | + |
All mutants were constructed and isolated previously (8, 9). In the mutants shown, DNA sequence letters in bold indicate changes from the wild-type, underlined letters indicate permanganate-modifiable thymine bases, and dotted lines correspond to deleted regions. In mutants +3, +6, and +11 the indicated sequence was inserted between the 13-mers. In mutants oriV3–6 and oriV3–11, the indicated sequence was inserted between the DnaA-box region and the AT-rich region (8). TF and FII results were presented previously (8, 9). FI*, KMnO4 and EM results are from this work except KMnO4 with oriV3–6 (8). TF, transformation frequency; FII, replication activity in E. coli crude extract; FI*, DnaB helicase-mediated template unwinding; KMnO4, local strand opening; EM, helicase loading to the AT-rich region; +, activity of the altered origin similar in comparison to the wild-type origin activity; +/−, lower activity; −, no (or substantially reduced) activity; nt, not tested.
Our results demonstrated that the lack of replication activity of the analyzed oriV minireplicons with altered 13-mer sequences could be a result of disturbances in helicase-dependent template unwinding. Because the formation of the FI* is a result of origin opening, helicase recruitment, and finally helicase complex activity, we could not exclude the possibility that the observed disturbances in helicase activity were due to failures at earlier steps of replication initiation, including open complex formation.
Open Complex Formation Is Affected by Sequence Changes Within the AT-Rich Region.
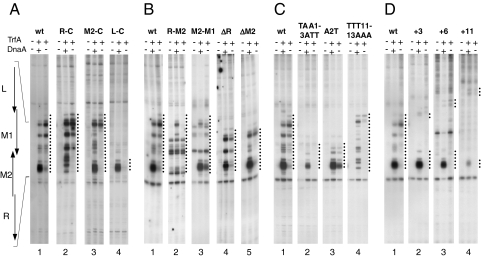
Full RK2 origin opening occurs in the presence of HU, TrfA, and DnaA proteins (3). To explore the possibility that mutations within the AT-rich region reduce DNA helix destabilization, we performed potassium permanganate (KMnO4) footprinting and compared the extent of origin opening for the wild-type and mutated oriV templates. KMnO4 reacts preferentially with pyrimidines in single-stranded DNA, modifying primarily thymines. Specific modification of plasmid supercoiled DNA can be detected by the termination of a primer extension reaction from a 32P-labeled primer at the modified residues. Because the majority of thymine residues present within the AT-rich region of RK2 origin are located on the top DNA strand, the primer extension analysis was carried out only on that strand. The pattern of KMnO4 modifications of the top strand of the plasmid RK2 wild-type origin is shown in Fig. 2. The footprinting experiments were carried out in the presence of either TrfA and DnaA protein or only TrfA. For each series (Fig. 2 A–D), wild-type template analysis was performed as a control.
Fig. 2.
Strand opening of mutant oriV templates. Potassium permanganate footprinting was carried out as described in Materials and Methods. TrfA and DnaA at 100 ng were present in the reactions as indicated. Dotted lines indicate the region where the DNA helix destabilization occurs. Position and orientation of the 13-mers for wild-type oriV is depicted on the left.
The experiments show that for six mutants (L-C, TAA1–3ATT, A2T, +3, +6, +11), the extent of opening was significantly reduced (Fig. 2 A, C, and D). For all these mutants, the DNA origin destabilization occurred only at the right 13-mer (R). Interestingly, the origin opening was reduced even in mutants with unchanged internal thermodynamic stability (TAA1–3ATT and A2T), indicating that this is not the only criterion for the functionality of AT-rich region. Reduction of the extent of origin opening was observed for two deletion mutants, ΔR and ΔM2 (Fig. 2B). The formation of full-length open complex that covers the whole AT-rich region (all four 13-mers) was observed for five mutants. Two of them, R-M2 and M2-M1, displayed an unusual feature. Partial origin destabilization appeared even in the absence of initiation proteins (Fig. 2B). For three other mutants R-C, M2-C, and TTT11–13AAA (Fig. 2 A and C), the extent of open complex formation was similar to that of the wild-type template. Similar to wild-type, in these mutants the open complex was formed also in the absence of DnaA protein, but it occurred at the left part of the AT-rich region (Fig. 2 A and C). Even though the extent of origin opening is the same for all these five mutants, a difference was observed in the pattern of permanganate modifications. This difference, however, can be attributed to alterations in nucleotide sequence, especially the thymine residues, of the analyzed mutants.
Our results demonstrate that the majority of the analyzed mutations within the AT-rich region have a significantly reduced plasmid open complex formation. This is similar to the situation in which introduced mutations within the E. coli oriC AT-rich region decreased or prevented the origin opening (11). Surprisingly, however, our analysis of oriV mutants (R-C, M2-C, R-M2, M2-M1 and TTT11–13AAA) revealed that the extent of origin opening was the same as for the wild-type template. This raised the possibility that low or absence of helicase activity of these mutants could be the result of disturbances of helicase complex loading at the open AT-rich region.
Mutations Within the 13-mers Affect Helicase Loading at the Open AT-Rich Region.
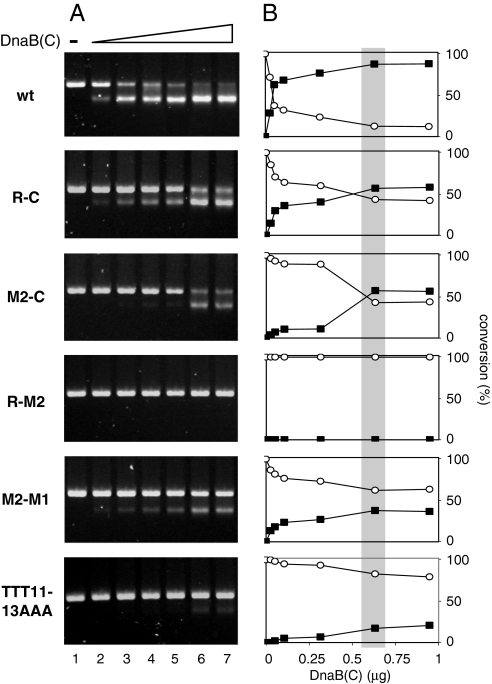
For the analysis of helicase complex formation at the RK2 origin, we used in vitro cross-linking followed by electron microscopy (EM) analysis (see Materials and Methods). This approach allowed identification of the position of the helicase complex on the replication origin. To identify the optimal helicase concentration for these experiments, we used various DnaBC concentrations to analyze the unwinding of the wild-type oriV template (Fig. 3 A and B). We determined a saturating helicase concentration (600 ng per reaction), in which complete conversion to the FI* form was observed. The complete conversion indicates that a vast majority of plasmid particles were successfully loaded by the helicase complex. When the oriV mutants with an unchanged origin opening but reduced helicase activity (R-C, M2-C, R-M2, M2-M1, and TTT11–13AAA) (see above) were analyzed, the highest level of FI* conversion was ≈50% (Fig. 3 A and B). Only very limited or no template unwinding activity was detected on the TTT11–13AAA and R-M2 mutant templates (Fig. 3 A and B). Despite a high molar excess (100-fold) of DnaBC in comparison to the supercoiled DNA template, no helicase activity was detected on the R-M2 template (Fig. 3A, lane 7).
Fig. 3.
Efficiency of unwinding by DnaB helicase at oriV mutant templates. The reactions were performed in the presence of E. coli DnaA and TrfA proteins as described in Materials and Methods. (A) (lanes 1–7): Reactions containing the increasing amounts of DnaBC helicase complex (0, 25, 50, 100, 300, 600, 900 ng). (B) Efficiency of conversion of supercoiled (FI) (opened circles) to the extensively unwound FI* DNA (filled squares), calculated on the basis of densitometry analysis. The gray bar indicates the saturating amount of DnaBC helicase complex in the reaction.
The above experiments allowed identification of the optimal DnaBC concentration (gray rectangle on Fig. 3B) to use for cross-linking and EM analysis (see below). Also, these results demonstrated that the inhibition of helicase unwinding activity as a result of alterations of 13-mers could not, or only to a limited extent, be compensated for by increasing the concentration of the DnaBC complex.
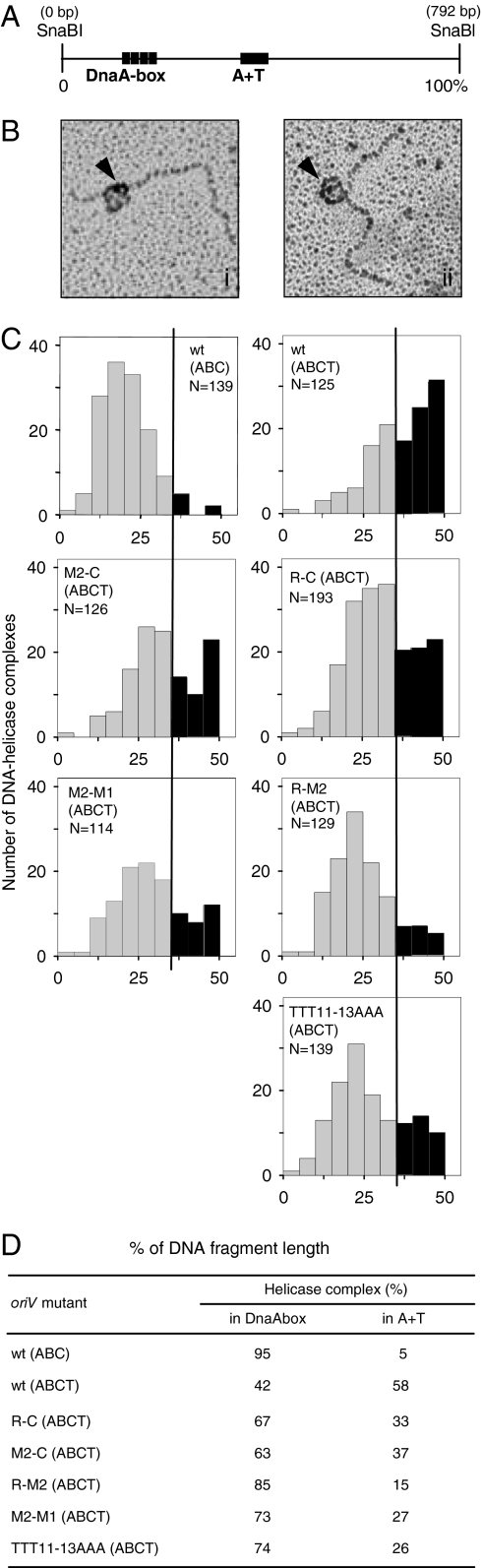
It has been shown that TrfA interacts with DnaB helicase recruited via interaction with DnaA protein at DnaA-boxes in oriV, located upstream of the AT-rich sequence (22). Therefore TrfA could possibly play a role in the translocation of the helicase complex from the DnaA-boxes to the AT-rich region. To explore this possibility, we assembled the reactions by using saturating concentrations of DnaBC as described above for FI* analysis. The utilization of the DnaB K236A (18) mutant protein, which forms nucleoprotein complexes but does not unwind DNA template, allowed for the analysis of the position of the helicase complex in the plasmid origin. After cross-linking with formaldehyde, the localization of the helicase complex on the 792 bp wild-type oriV fragment (Fig. 4A) was determined (see Materials and Methods). In a control reaction, the formation of the nucleoprotein complex containing DnaA, DnaB, and DnaC proteins, but not TrfA, was analyzed. Statistical analysis confirmed our previously published observation (22) that the majority of DnaB helicase (95%) was found at the DnaA-boxes (Fig. 4 B(i), C, and D). Only in 5% of the analyzed DNA particles was DnaB detected within the AT-rich region. When TrfA protein was added to the reaction mixture, the proportion changed significantly. In 58% of the plasmid particles, helicase was detected in the oriV fragment corresponding to the AT-rich region (Fig. 4 B(ii), C, and D). This result demonstrates translocation of the helicase complex from the DnaA-boxes to the AT-rich region. Because the addition of TrfA results in the origin opening (ref. 3 and above) and the release of helicase activity (ref. 23 and above), we concluded that the observed DnaB complex was most probably formed at the open origin structure. After the addition of TrfA, the number of helicase complexes at the DnaA-box region decreased substantially (Fig. 4). Because TrfA interaction with DnaB helicase has been reported (22) and it also has been shown that TrfA interacts with the helicase complex formed on the DnaA-boxes (22), we propose direct involvement of TrfA in the observed helicase complex translocation. The interaction of E. coli DnaB helicase with other plasmid initiating proteins has been demonstrated for plasmids R6K (24) and pSC101 (25).
Fig. 4.
Effect of 13-mers mutations on DnaB complex formation at the oriV AT-rich region. To analyze the position of the helicase complex at the oriV origin, we performed cross-linking experiments and EM analysis as described in Materials and Methods. Reactions contained wild-type or mutant oriV templates; HU; DnaA; DnaB; DnaC; and, if indicated, TrfA. (A) Schematic depiction of an oriV SnaBI, 792-bp fragment with marked DnaA-boxes and AT-rich region. (B) Typical EM images of DnaB helicase (indicated with an arrow) at the DnaA-box sequences of oriV (i) or at the AT-rich region (ii). (C) Histograms present measurements of the helicase complex position on the analyzed DNA fragments. The black line indicates the midpoint between the DnaA-boxes and the AT-rich region. The helicase complexes at these regions are indicated by light gray and dark gray, respectively. N indicates the number of particles taken for the EM analysis. Statistical analysis of the obtained results is given in the table (D).
To complete our analysis, we tested the possibility that the alterations within the oriV AT-rich region reduced the efficiency of helicase loading at the open origin structure. We tested assembly of the helicase on oriV templates with 13-mer alterations (R-C, M2-C, R-M2, M2-M1, and TTT11–13AAA) that, as demonstrated above, do not affect origin opening but diminish helicase activity. In comparison to the wild-type template, assembly of the helicase complex at the AT-rich region was substantially reduced in all analyzed mutants (Fig. 4 C and D). The reactions were carried out in the presence of TrfA and under the same experimental conditions as for the wild-type plasmid. The lowest percentage (15%) of DnaB molecules in the AT-rich region was observed for mutant R-M2. Although we did not observe any specific pattern of 13-mers alterations affecting helicase loading, the reduced helicase complex formation was observed on templates, with changes introduced into the 13-mer M2 or R located in the right part of the oriV AT-rich region (Table 1). The reduction of helicase complex formation at the AT-rich region of the mutant origins generally corresponded to the reduction of helicase activity observed on the same templates (Figs. 3 and 4D); however, for two mutants (R-M2 and TTT11–13AAA) the result was not so striking. Cross-linking and utilization of the DnaB K236A mutant could account for the stabilization of helicase complexes at those templates.
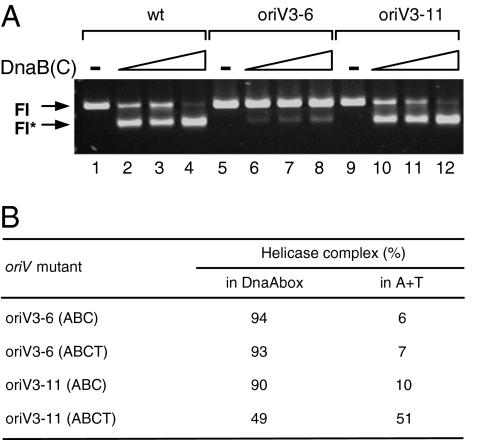
Helicase Translocation Requires Appropriate Facing of the Origin Motifs.
Our EM studies showed that the position of the helicase on the DNA template changed as a result of the addition of TrfA, a process we defined as translocation. However, it hadn't been directly shown whether it was an actual change in the helicase position on the DNA or a change in the specific site on the DNA template that was in direct contact with the helicase. Several possible mechanisms behind the observed process could be considered. To address this issue, we conducted experiments by using oriV mutants that contained insertions between the DnaA-box region and the AT-rich region. The insertion of 6 bp (oriV3–6 mutant) knocked out template activity in vivo (8), which we identified as not due to an inhibition of origin opening (8), but as the result of the failure of the helicase to translocate and a consequent loss of helicase activity (Fig. 5 A and B). In contrast to the 6-bp insertion, the introduction of an approximately full helical turn (oriV3–11 mutant) between the DnaA-box region and the AT-rich region allowed helicase translocation and activity in vitro (Fig. 5 A and B) and replication activity in vivo (8). These results demonstrate that translocation of helicase from the DnaA-boxes to the 13-mers requires appropriate facing of the origin motifs. This implies that helicase translocation is not a spontaneous dissociation from its interaction with DnaA and DnaA-boxes and establishment of contact with ssDNA within the 13-mers. Also, it is very unlikely that the helicase slides from one position, the DnaA-boxes, to another and thereby establishes contact with the opened AT-rich region. Obviously, helicase translocation requires some form of face-to-face contact which most probably could be promoted by a specific change of the origin DNA structure. Possibly, TrfA together with HU changes the oriV nucleoprotein structure in such a way that helicase contact with the ssDNA within the 13-mers is favored, resulting in the release of helicase from the DnaA-boxes. This mechanism would require TrfA interaction with DnaB (22) and TrfA cooperation with the host DnaA protein, which recruits the helicase to DnaA-boxes (23) and is involved in stabilization of origin melting by TrfA (3). The involvement of histone-like proteins in this process is essential, as HU is required for origin opening by TrfA (3). Also, TrfA, in an iteron- and/or 13-mer-dependent manner, could possibly form filaments that restructure the DNA at the replication origin, promoting both origin opening and helicase translocation. It was recently proposed for plasmid pPS10 RepA protein that defined DNA sequences promote the assembly of the protein into a distinct amyloid nanostructure (26). TrfA, similar to pPS10 RepA, contains regions that form filaments through β-sheets, which we identified by using the TANGO algorithm (http://tango.crg.es).
Fig. 5.
Helicase translocation requires appropriate facing of the origin motifs. (A) FI* reactions containing increasing amounts of the DnaBC helicase complex (at 0, 100, 300, 600 ng) on wild-type or mutant templates containing insertions of 6 bp (oriV3–6) or 11 bp (oriV3–11) between the DnaA-box region and the AT-rich region containing 13-mers. (B) Results obtained in EM experiments performed for those mutants as described in Materials and Methods.
The requirement for a specific sequence for helicase activity within the AT-rich region could be a general feature of different replicons. A selective activation by stretches of thymine residues of the Mcm4/6/7 helicase complex has been reported and proposed to be a determinant for the selection of DNA replication initiation sites in mammalian genomes (27). We demonstrate that loading of the DnaB helicase, primarily recruited by host DnaA protein at DnaA-boxes, depends on the activity of the RK2 plasmid replication initiator to facilitate helicase translocation to the AT-rich opened region, in a reaction requiring specific sequence of the 13-mers and appropriate facing of the origin motifs.
Materials and Methods
Plasmids, Proteins, and Reagents.
The plasmids containing mutations within the AT-rich region were constructed as described previously (5). Plasmids oriV3–6 and oriV3–11 are described in Doran et al. (8). For TrfA, preparations of the histidine-tagged version of the largely monomeric TrfA mutant protein His-6-TrfA G254D/S267L, which exhibits a high specific activity for iteron binding (28), was used. E. coli DnaA, wild-type DnaB, the K236A ATP-non hydrolyzing mutant DnaB (18), and E. coli DnaC proteins were purified according to published procedures (29–31). Commercially available proteins and chemicals were as follows: SSB and T4 polynucleotide kinase from Promega; BSA (fraction V), creatine phosphate, creatine kinase, rNTPs, KMnO4, and Protein A 10-nm colloidal gold labeled for EM analysis from Sigma; Sepharose CL-4B from Amersham Pharmacia; and the MiniPrep DNA purification kit from A&A Biotechnology. AmpliTaq Gold polymerase and [32P]ATP were from Perkin-Elmer. A 24-mer (GCTTGCATGCCTGCAGGTCGACTC) used for primer extension was from Thermohybaid.
Permanganate Footprinting and Primer Extension.
Permanganate footprinting was carried out as described (3), with modifications. The standard reaction mixture (25 μl) contained 8 ng of HU, 100 ng of E. coli DnaA, 300 ng of oriV DNA, and 100 ng of His-6-TrfA G254D/S267L. Each sample was purified further by using a Gene Clean III kit (BIO101). The analysis was performed on the top DNA strand with the 24-nucleotide primer, as described previously (3).
Helicase Activity Assay (FI*).
The reactions were performed essentially as described (23). Wild-type and mutated RK2 supercoiled minireplicons (300 ng) were used as the DNA template. Unless indicated otherwise, the mixture contained DnaA (10 ng), DnaB (600 ng), DnaC (120 ng), His-6-TrfA G254D/S267L protein (100 ng), HU (5 ng), SSB (230 ng), and gyrase (120 ng). The samples were electrophoresed at 25 V for 22 h, and the gel was stained with ethidium bromide. The helicase activity was calculated by densitometry, using BioRad Quantity One software. pUC19A supercoiled DNA (1000 ng) containing cluster of oriV DnaA-boxes was used in the experiments testing helicase loading in trans.
Cross-Linking and Electron Microscopy.
The reactions were prepared similarly as described (22), in the buffer (100 μl) containing supercoiled DNA templates (1,200 ng), and proteins added in the following order: E. coli HU (20 ng), DnaA (60 ng), DnaB K236A (2,400 ng), DnaC (480 ng), and TrfA 33kDa (400 ng). Samples were cross-linked by the addition of 0.1% formaldehyde. SnaBI restriction digestion, followed by gel filtration chromatography on Sepharose CL-4B, was performed to obtain a 792-bp oriV fragment containing DnaA-boxes and the AT-rich region. Samples were adsorbed on mica sheets, stained with uranyl acetate, platinum-carbon coated, and further examined at 80 kV in a Philips CM100 electron microscope. Images were statistically analyzed for the position of the helicase complex by using AnalySIS software.
Acknowledgments.
We thank Dr. Aresa Toukdarian for critical reading of the manuscript. This work was supported by the Polish Ministry of Science and Higher Education Grant 2P04A02730, The Foundation for Polish Science, and the EMBO/HHMI YIP Program. M.R. and L.K. are funded by The Foundation for Polish Science.
Footnotes
The authors declare no conflict of interest.
References
- 1.Stalker DM, Thomas CM, Helinski DR. Nucleotide sequence of the region of the origin of replication of the broad host range plasmid RK2. Mol Gen Gen. 1981;181:8–12. doi: 10.1007/BF00338997. [DOI] [PubMed] [Google Scholar]
- 2.Perri S, Helinski DR, Toukdarian A. Interactions of plasmid-encoded replication initiation proteins with the origin of DNA replication in the broad host range plasmid RK2. J Biol Chem. 1991;266:12536–12543. [PubMed] [Google Scholar]
- 3.Konieczny I, Doran KS, Helinski DR, Blasina A. Role of TrfA and DnaA proteins in origin opening during initiation of DNA replication of the broad host range plasmid RK2. J Biol Chem. 1997;272:20173–20178. doi: 10.1074/jbc.272.32.20173. [DOI] [PubMed] [Google Scholar]
- 4.Bramhill D, Kornberg A. Duplex opening by DnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988;52:743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- 5.Kowalski D, Eddy MJ. The DNA unwinding element: A novel, cis-acting component that facilitates opening of the Escherichia coli replication origin. EMBO J. 1989;8:4335–4344. doi: 10.1002/j.1460-2075.1989.tb08620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gille H, Messer W. Localized DNA melting and structural pertubations in the origin of replication, oriC, of Escherichia coli in vitro and in vivo. EMBO J. 1991;10:1579–1584. doi: 10.1002/j.1460-2075.1991.tb07678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hwang DS, Kornberg A. Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J Biol Chem. 1992;267:23083–23086. [PubMed] [Google Scholar]
- 8.Doran KS, Konieczny I, Helinski DR. Replication origin of the broad host range plasmid RK2. Positioning of various motifs is critical for initiation of replication. J Biol Chem. 1998;273:8447–8453. doi: 10.1074/jbc.273.14.8447. [DOI] [PubMed] [Google Scholar]
- 9.Kowalczyk L, Rajewska M, Konieczny I. Positioning and the specific sequence of each 13-mer motif are critical for activity of the plasmid RK2 replication origin. Mol Microbiol. 2005;57:1439–1449. doi: 10.1111/j.1365-2958.2005.04770.x. [DOI] [PubMed] [Google Scholar]
- 10.Hsu J, Bramhill D, Thompson CM. Open complex formation by DnaA initiator protein at the Escherichia coli chromosomal origin requires the 13-mer precisely spaced relative to the 9-mers. Mol Microbiol. 1994;11:903–911. doi: 10.1111/j.1365-2958.1994.tb00369.x. [DOI] [PubMed] [Google Scholar]
- 11.Hwang DS, Kornberg A. Opposed actions of regulatory proteins, DnaA and IciA, in opening the replication origin of Escherichia coli. J Biol Chem. 1992;267:23087–23091. [PubMed] [Google Scholar]
- 12.Slater S, et al. E. coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in DNA replication initiation and origin sequestration. Cell. 1995;82:927–936. doi: 10.1016/0092-8674(95)90272-4. [DOI] [PubMed] [Google Scholar]
- 13.Brendler T, Abeles A, Austin S. A protein that binds to the P1 origin core and the oriC 13mer region in a methylation-specific fashion is the product of the host seqA gene. EMBO J. 1995;14:4083–4089. doi: 10.1002/j.1460-2075.1995.tb00080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuller RS, Funnell BE, Kornberg A. The dnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell. 1984;38:889–900. doi: 10.1016/0092-8674(84)90284-8. [DOI] [PubMed] [Google Scholar]
- 15.Speck C, Messer W. Mechanism of origin unwinding: Sequential binding of DnaA to double- and single-stranded DNA. EMBO J. 2001;20:1469–1476. doi: 10.1093/emboj/20.6.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Learn BA, Um S-J, Huang L, McMacken R. Cryptic single-stranded-DNA binding activities of the phage lambda P and Escherichia coli DnaC replication initiation proteins facilitate the transfer of E. coli DnaB helicase onto DNA. Proc Natl Acad Sci USA. 1997;94:1154–1159. doi: 10.1073/pnas.94.4.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davey MJ, Fang L, McInerney P, Georgescu RE, O'Donnell M. The DnaC helicase loader is a dual ATP/ADP switch protein. EMBO J. 2002;17:3148–3159. doi: 10.1093/emboj/cdf308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang L, Davey MJ, O'Donnell M. Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol Cell. 1999;4:541–553. doi: 10.1016/s1097-2765(00)80205-1. [DOI] [PubMed] [Google Scholar]
- 19.Marszalek J, Kaguni JM. DnaA protein directs the binding of DnaB protein in initiation of DNA replication in Escherichia coli. J Biol Chem. 1994;269:4883–4890. [PubMed] [Google Scholar]
- 20.Weigel C, Seitz H. Strand-specific loading of DnaB helicase by DnaA to a substrate mimicking unwound oriC. Mol Microbiol. 2002;46:1149–1156. doi: 10.1046/j.1365-2958.2002.03232.x. [DOI] [PubMed] [Google Scholar]
- 21.Ozaki S, et al. A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J Biol Chem. 2008;283:8351–8362. doi: 10.1074/jbc.M708684200. [DOI] [PubMed] [Google Scholar]
- 22.Pacek M, Konopa G, Konieczny I. DnaA box sequences as the site for helicase delivery during plasmid RK2 replication initiation in Escherichia coli. J Biol Chem. 2001;276:23639–23644. doi: 10.1074/jbc.M100255200. [DOI] [PubMed] [Google Scholar]
- 23.Konieczny I, Helinski DR. Helicase delivery and activation by DnaA and TrfA proteins during the initiation of replication of the broad host range plasmid RK2. J Biol Chem. 1997;272:33312–33318. doi: 10.1074/jbc.272.52.33312. [DOI] [PubMed] [Google Scholar]
- 24.Ratnakar PV, Mohanty BK, Lobert M, Bastia D. The replication initiator protein pi of the plasmid R6K specifically interacts with the host-encoded helicase DnaB. Proc Natl Acad Sci USA. 1996;93:5522–5526. doi: 10.1073/pnas.93.11.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Datta HJ, Khatri GS, Bastia D. Mechanism of recruitment of DnaB helicase to the replication origin of the plasmid pSC101. Proc Natl Acad Sci USA. 1999;96:73–78. doi: 10.1073/pnas.96.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giraldo R. Defined DNA sequences promote the assembly of a bacterial protein into distinct amyloid nanostructures. Proc Natl Acad Sci USA. 2007;104:17388–17393. doi: 10.1073/pnas.0702006104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.You Z, et al. Thymine-rich single-stranded DNA activates Mcm4/6/7 helicase on Y-fork and bubble-like substrates. EMBO J. 2003;22:6148–6160. doi: 10.1093/emboj/cdg576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blasina A, Kittell BL, Toukdarian AE, Helinski DR. Copy-up mutants of the plasmid RK2 replication initiation protein are defective in coupling RK2 replication origins. Proc Natl Acad Sci USA. 1996;93:3559–3564. doi: 10.1073/pnas.93.8.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caspi R, et al. A broad host range replicon with different requirements for replication initiation in three bacterial species. EMBO J. 2001;20:3262–3271. doi: 10.1093/emboj/20.12.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caspi R, Helinski DR, Pacek M, Konieczny I. Interactions of DnaA proteins from distantly related bacteria with the replication origin of the broad host range plasmid RK2. J Biol Chem. 2000;275:18454–18461. doi: 10.1074/jbc.M000552200. [DOI] [PubMed] [Google Scholar]
- 31.SanMartin MC, Stamford NPJ, Dammerova N, Dixon NE, Carazo JM. A structural model for the Escherichia coli DnaB helicase based on electron microscopy data. J Struct Biol. 1995;114:167–176. doi: 10.1006/jsbi.1995.1016. [DOI] [PubMed] [Google Scholar]