Abstract
Organ shape and size, and, ultimately, organ function, relate in part to the cell and tissue spatial arrangement that takes place during embryonic development. Despite great advances in the genetic regulatory networks responsible for tissue and organ development, it is not yet clearly understood how specific gene functions are linked to the specific morphogenetic processes underlying the internal organ asymmetries found in vertebrate animals. During female chick embryogenesis, and in contrast to males where both testes develop symmetrically, asymmetrical gonad morphogenesis results in only one functional ovary. The disposition of paired organs along the left–right body axis has been shown to be regulated by the activity of the homeobox containing gene pitx2. We have found that pitx2 regulates cell adhesion, affinity, and cell recognition events in the developing gonad primordium epithelia. This in turn not only allows for proper somatic development of the gonad cortex but also permits the proliferation and differentiation of primordial germ cells. We illustrate how Pitx2 activity directs asymmetrical gonad morphogenesis by controlling mitotic spindle orientation of the developing gonad cortex and how, by modulating cyclinD1 expression during asymmetric ovarian development, Pitx2 appears to control gonad organ size. All together our observations indicate that the effects elicited by Pitx2 during the development of the female chick ovary are critical for cell topology, growth, fate, and ultimately organ morphogenesis and function.
Keywords: cyclin D1, left–right asymmetry, chick ovary development, mitotic orientation, primordial germ cell
Despite the initial bilateral symmetry of the vertebrate body plan, internal organs, such as the heart, stomach, and intestines, all have a characteristic asymmetric structure and are asymmetrically positioned in the body cavity. The acquisition of proper left–right asymmetries is achieved in a highly conserved manner during embryo development. Since the seminal work more than a decade ago that identified a functional link between asymmetrical gene expression and organ asymmetry (1), our understanding of the molecular and cellular events involved in the establishment of left–right asymmetry in vertebrates has increased enormously. This is indeed the case in the initial establishment of left–right asymmetrical differences that occur during gastrulation as well as during the immediately subsequent stages of embryo development when the local asymmetrical cues in and around the node are deployed to broader domains of gene expression in the lateral plate mesoderm. Once side-specific gene expression domains are established and stabilized in the lateral plate mesoderm, left–right information is transferred to the organ primordial, where left and right side-specific morphogenetic programs are executed (reviewed in refs. 2–6).
The identification of several genes that display side-specific patterns of expression within the developing organs has provided an entry point for understanding the molecular and cellular mechanisms underlying asymmetric morphogenesis. These genes include, among others, the bicoid-type homeobox gene pitx2. Unlike other asymmetric genes, pitx2 expression is maintained during the development of several organ primordia, making it a good candidate to be involved in both mediating the maintenance of left–right information from the lateral plate mesoderm to its derived developing organs and executing it at a cellular level. Indeed, Pitx2 loss- and gain-of-function experiments can cause organ-specific laterality defects in all vertebrates analyzed to date (6–21).
Notwithstanding these advances, we are still far away from understanding how organ primordia interpret left–right genetic information during organogenesis, i.e., how broad domains of side-specific gene expression in the lateral plate mesoderm translate to asymmetric organ development. Moreover, our knowledge on how asymmetric gene activity is linked to specific cellular processes is also still scarce.
Here, and by focusing on the development of the female chick gonads, we try to gain insights on how local processes involving differential control of cell proliferation and/or cell death, as well as instructive changes in cell fate, are likely to be implicated in providing morphogenetic cues that in turn direct asymmetrical organ development.
Results and Discussion
Subcellular Asymmetries of the Left Versus Right Female Chick Gonad.
In most vertebrates, the urogenital system develops in a symmetrical, bilateral pattern in both sexes. In birds, however (22), bilateral development of the paired gonads is sex-dependent. During female chick embryogenesis, and in contrast to males, where both testes develop symmetrically, asymmetrical gonad morphogenesis results in only one functional ovary, the left one; the right gonad, initially morphologically indistinguishable from the left one, degenerates at later stages of development.
The gonad primordium arises early in embryogenesis from a thickening of the ventromedial surface of the mesonephros located between somites 18 and 22 at stages 19–20 HH (23). This initial ridge, which will constitute the somatic compartment of the gonad, is composed of an outer epithelial cell layer (cortex) and the underlying medulla. Primary (in the medulla) and secondary (in the cortex) cords are formed and subsequently colonized by the primordial germ cells coming from the germinal crescent via the vascular system (24, 25). At this stage, the left and right primitive gonads are morphologically indistinguishable in both male and female embryos (26). The anatomical asymmetry displayed specifically by female embryos does not become apparent until stages 29–30 HH and is macroscopically evident 2–3 days later (stages 34–36 HH). At these stages, the future left ovary displays a thickened differentiated surface epithelia (this is the location where future folliculogenesis will take place). However, in the right gonad, which is significantly smaller at this time than the left one, the left-sided characteristic differentiation and thickening of the epithelia have not yet occurred, causing the gonad primordia to degenerate.
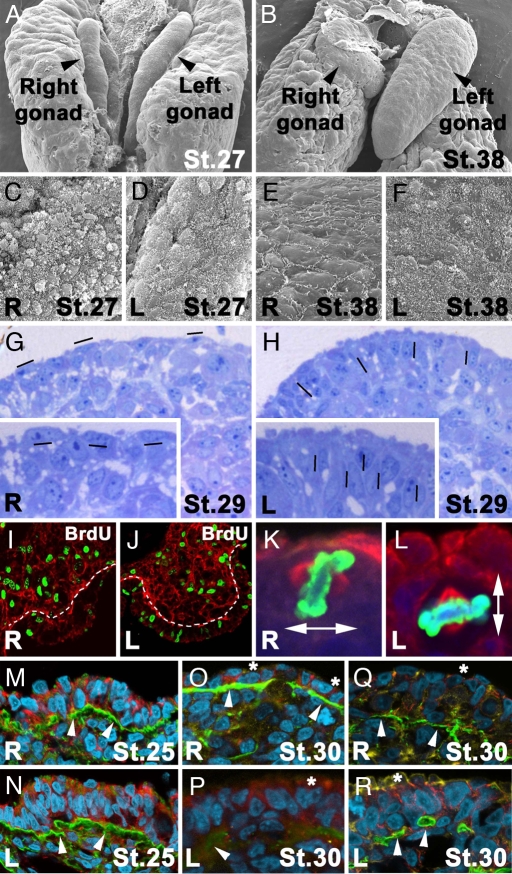
The overt morphological asymmetries of the left and right gonads observed at stages 30 HH and beyond are preceded by differences in the spatial and structural disposition of the left and right epithelia (Fig. 1 A and B). Until stage 27 HH, the gonad's outer cells from both the left and right epithelia are globular in shape and completely covered by pilli that consist of piled-shape structures (Fig. 1 C and D). Subsequently, cells from the right epithelia start to lose their globular shape, and their pilli adopt a flattened morphology and become restricted to cell junctions (Fig. 1E, compare with F). Histological sections reveal differences in the compactness of the nuclei between the left and right epithelia, as well as differences in their spatial orientation. Most of the nuclei in the right gonad orient their longest axis parallel to the cortex, whereas the left gonadal nuclei are oriented perpendicular to the epithelial surface (Fig. 1 G and H). Concomitantly to the changes described in the spatial arrangement of cells within the gonad epithelia, BrdU incorporation experiments demonstrate differences in cell proliferation rates between the right and left epithelia from stage 27 to stage 29 HH. The variations described in Fig. 1 I and J are restricted to the cortex because no significant differences are observed in the medullae.
Fig. 1.
Asymmetrical differences in female chick gonad embryogenesis. (A and B) Scanning electron micrographs at stage 27 (A) and stage 38 (B) HH. (C–F) Right (C and E) and left (D and F) surface of symmetrical stage 27 HH (C and D) and asymmetrical stage 38 HH (E and F) gonads. Note the loss of a globular shape and pilli distribution on the right gonad (C and E). (G and H) Histological sections of right (G) and left (H) gonads at stage 29 HH stained with Toluidine blue illustrating condense packaging of nuclei in the left gonad cortex (H). Magnifications of right and left cortex are evident in the Insets of G and H. Hyphens show the orientation of oval nuclei. (I and J) BrdU incorporation (green) in right (I) and left (J) gonad nuclei after a 1-h pulse at stage 28 HH. A white line marks the borders of the gonad cortex to compare differential proliferation. Basal lamina is evident by laminin immunostaining in red. (K and L) Representative mitotic cells for right (K) and left (L) gonads with perpendicular and parallel metaphase planes, respectively, at stage 27 HH (nuclei shown in blue, Phospho Histone H3 in green, and α-tubulin in red). (M–P) Immunohistochemistry detection for laminin (green) and β-catenin (red) at stage 25 HH (M and N) and 30 HH (O and P) in the right (M and O) and left (N and P) gonads. (Q and R) Immunostaining for laminin (green) and N-cadherin (red) at stage 30 HH in the right (Q) and left (R) gonads. In M–R, DNA is shown in cyan and actin is shown in yellow. At stage 30 HH, basal lamina continuity is maintained in the right gonad (O and Q) but not in the left gonad (P and R), compared with the symmetry detected in both gonads at stage 25 (M and N; white arrowheads in M–R point to basal lamina). Note a loss of the intense detection of β-catenin and N-cadherin in the apical cytocortex at stage 30 HH (asterisks) when morphological asymmetry between left and right gonads is evident (O–R).
Correct cell rate proliferation and proper spatial arrangement are crucial for cell–cell interactions and, consequently, for the transmission of inductive signals between cells, which, in turn, facilitate the correct progression of organ development. Adhesion molecules and extracellular matrix components are known to play critical roles during cell–cell interactions (reviewed in refs. 27 and 28). Immunohistochemistry studies reveal dynamic changes in extracellular and subcellular components during development of the left and right gonads (Fig. 1 K–R). Before any overt morphological changes are detected (i.e., before stages 26–27 HH) cell nuclei from both the right and left female gonads are organized in a similar spatial orientation, with a continuous basal lamina underlying the cortex in both gonads and without obvious differences in the distribution of N-cadherin, β-catenin, or actin between the left and right gonads (Fig. 1 M and N). From stage 28 HH onwards, differences between the left and right gonads start to be observed (Fig. 1 O–R). Although basal lamina structure on the right gonad is not completely maintained, the limits of the right cortex are clearly seen (Fig. 1 O and Q). In the case of the left gonad, basal lamina continuity is lost and only fragments of it are visible (Fig. 1 P and R). Subcellular localization of N-cadherin, β-catenin, and actin in the right gonad is detected only laterally to the nuclei in the portion of the cytoplasm that localizes close to cell junctions (Fig. 1 O and Q). On the contrary, in the left gonad, N-cadherin, β-catenin, and actin localize laterally to the nuclei, in the external cytoplasmic part of the cortex (Fig. 1 P and R).
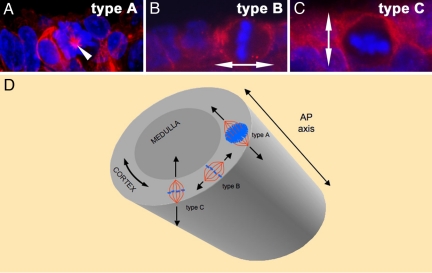
The above-described differences between the left and right gonads are accompanied by differences in the orientation plane of cell division at stage 27 HH [Fig. 1 K and L and statistical analysis in supporting information (SI) Table S1]. For simplification we have classified the cells undergoing mitosis in the cortex into three different types (summarized in Fig. 2). Type A mitosis contains cells whose metaphase plane is oriented in an orthogonal way when referred to the anteroposterior (AP) axis of the gonad (Fig. 2A). Type B refers to dividing cells in which the metaphase plate is parallel to the AP axis and perpendicular to the cortex (Figs. 1K, 2B, and 3S). Finally, type C mitosis includes cells with a mitotic plate parallel to the AP axis and to the cortex (Figs. 1L, 2C, and 3 T and U). The plane of cytokinesis in the majority of cells undergoing mitosis in the right cortex is perpendicular to the cortex plane or to the AP axis (type A and B mitosis in Fig. 2). However, in the left gonad, the plane of cytokinesis in most of the cells has a defined bias toward a 5parallel orientation with respect to the cortex plane (Fig. 1K). In fact, the distribution of the different types of mitotic divisions is significantly different when left and right cortex are compared (statistical analysis in Table S1). The main evident difference is the distribution of type C divisions (Figs. 1L and 2C), which is two times higher in the left cortex than in the right one (Figs. 1 K and L and 2 and Table S1). Overall, the differences in cell adhesion, affinity, and cell recognition of the left and right gonads' epithelia reported here may correlate with the correct orientation of mitotic divisions, which, in turn, have been shown in other contexts to play a crucial role in cell topology, growth, fate, and, ultimately, organ morphogenesis and function (29, 30).
Fig. 2.
Types of mitotic orientations in developing chick gonads. For simplification, mitotic orientations have been divided into three different groups, depending on the angle of the metaphase plate with respect to the AP axis of the gonad or the cortex. By abstraction, we can equate the gonadal cortex to a cylinder whose long axis aligns with the embryonic AP axis (D). Perpendicular A/P (type A): Mitotic cells whose metaphase plane has an orthogonal orientation with respect to the AP axis (mitosis type A in D). In transverse sections, the metaphase plate of this group of cells can be observed in its whole extension with almost all of the chromosomes in the same plane (A, white arrowhead points to the mitotic spindle that is oriented perpendicular to the plane of the image). Perpendicular to cortex (type B): The metaphase plate of these cells is located in a plane parallel to the AP axis (mitosis type B in D) and perpendicular to the cortex (type B in B). Double arrow shows orientation of mitotic spindle. Parallel to cortex (type C): In this group of cells, the metaphase plate also contains the AP axis (mitosis type C in D) and is oriented parallel to the cortex plane (type C in C). The double arrow shows the orientation of the mitotic spindle.
Fig. 3.
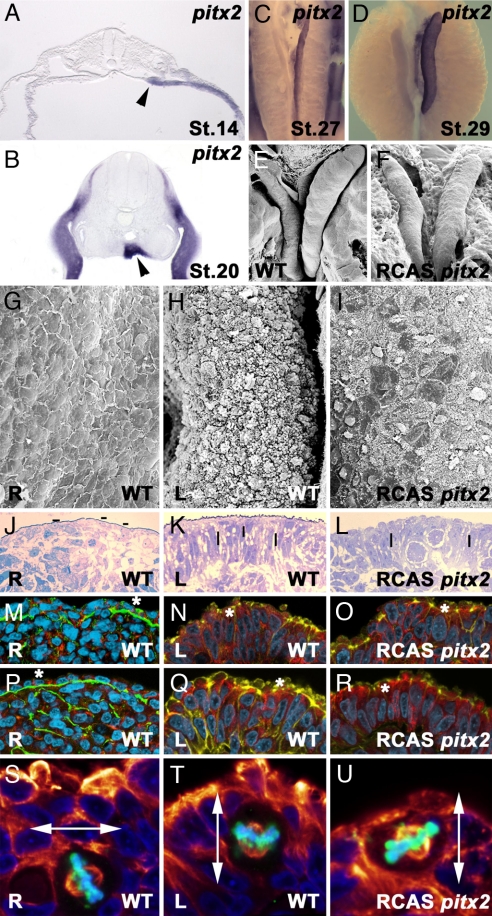
Pitx2c regulates gonad morphogenesis. (A and B) Histological sections showing pitx2c expression in left gonad primordia (black arrowhead) at stage 14 HH (A) and 20 HH (B). Left-sided pitx2c expression is detected before any overt difference is observed between the left and right gonads. Subsequently, expression is maintained through cortex asymmetric differentiation stages (stage 27 in C and stage 29 in D). Overgrowth of the right gonad 48 h after pitx2c overexpression (F) can be compared with a control stage 31 HH right gonad (E). (G–U) Phenotype of right gonads after pitx2c overexpression. (G, J, M, P, and S) Right gonads. (H, K, N, Q, and T) Left gonads. (I, L, O, R, and U) Right gonads after pitx2c overexpression. Scanning electron microscopy micrographs show patches with high pilli density in right gonads after pitx2c overexpression as observed for left wild-type gonads (I, compare with G and H). Histological sections show compactness of nuclei density and a shift to a perpendicular orientation on the right gonad after pitx2c overexpression (L, compared with J and K). Hyphens indicate nuclei orientation. Immunohistological sections show transformation of the right gonad to left phenotype after pitx2c overexpression with laminin and β-catenin (O, compare with M and N) and laminin and N-cadherin (R, compare with P and Q). Note the loss of basal lamina continuity, increase of nuclei packaging, and detection of N-cadherin, β-catenin, and actin in the external section of the cytoplasm after pitx2c overexpression (asterisks). In M–R, DNA is shown in cyan and actin is shown in yellow. (S–U) Cell division plane in pitx2c transformed (U) and in control (A/P-infected) right (S) and left (T) gonads (nuclei shown in blue, Phospho Histone H3 in green, and α-tubulin in red). pitx2c overexpression increases the percentage of parallel mitotic divisions in the right gonad. Double arrows point to the orientation of the mitotic spindle.
Pitx2 Activity Directs Asymmetric Female Gonad Development.
The bicoid type homeobox gene pitx2 is expressed in the left side of several organ primordia and has an instructive role in mediating left–right organ asymmetries (6–21). During female chick gonad development, pitx2c mRNA transcripts are restricted to the presumptive left gonad cortex epithelia at stage 14 HH (Fig. 3A) and subsequently to the left gonad cortex until the last stage examined (38 HH; Fig. 3 B–D and data not shown). No expression is detected on the right side at any stage of gonad development.
To study the effects that the presence of pitx2c may have on the right gonad we injected chick embryos, from stages 10–20 HH, in the right urogenital ridge with a retroviral vector containing pitx2c (8), the isoform specifically expressed in the left gonad. To assay the reliability and extension of in ovo infection, a replication-competent retrovirus expression GFP was injected under identical conditions and viral infection was visualized at later stages of development. Scanning electron microscopy micrographs show that patches of cells in the pitx2c-infected right cortex do not lose their globular surface and pilli distribution as control (infected with a retrovirus containing alkaline phosphatase) right gonads do after stage 30 HH (n = 90, 38%; Fig. 3I, compare with G and H). Moreover, histological sections show that nuclei distribution in the pitx2c-infected right cortex is compacted, their orientation is perpendicular to the cortex surface, and basal lamina continuity is lost as observed in control left gonads (Fig. 3 J–L). These structural changes are also accompanied by modifications in N-cadherin, β-catenin, and actin distribution (Fig. 3 M–R). These alterations can be specifically observed in the apical cytocortex, which is linked to the patches of cells with globular shape and higher densities of pilli (n = 260, 37%; Fig. 3 O and R; compare with M and N and with P and Q, respectively). Overall, the ectopic expression of pitx2c on the right cortex elicits cellular and subcellular changes that result in a phenotype resembling that of the left gonad.
Furthermore, analysis of cell division and spindle orientation reveals that whereas in the control right gonad most of the mitotic cells display an angle of division perpendicular to the cortex plane or perpendicular to the AP axis (type A and type B mitosis; Fig. 2 A and B and Table S1), forced expression of pitx2c alters the plane of mitotic division (type C mitosis; Fig. 2C and Table S1). This shift in the relative percentage of the different types of mitosis mirrors the higher amount of mitotic cells with parallel orientation observed in the control left cortex (Figs. 2 and 3 S–U and Table S1). The P value for the whole statistical analysis is <0.001.
The alterations elicited by forced expression of pitx2c are not restricted to the early stages of right gonad development because they are also reflected in the subsequent gonad growth dynamics and final gonad morphology. Indeed, forced expression of pitx2c erases the morphological asymmetries present during normal development between the left and right gonads (Fig. 3 G–R). Time course studies indicate that the normal regression that starts to be detected in the right gonad after stage 30 HH does not occur after ectopic expression of pitx2c, treatment that leads to a larger gonad size with a growth rate that resembles that of the left gonad (Fig. 3F, compare with E).
Overall, all of the observations above reported indicate that the forced expression of pitx2c on the right genital ridge is sufficient to rescue its programmed degeneration. Pitx2c is able to induce left isomerism and cortical differentiation (see also below and ref. 24). Thus, the asymmetric expression of pitx2c in the female left genital ridge during embryogenesis has an instructive role in eliciting cellular and subcellular behaviors that determine asymmetrical ovarian morphogenesis. Moreover, Pitx2c controls the major driving force for asymmetrical gonad morphogenesis that seems to reside in the differential distribution of mitotic orientation within the cortex. In fact, induction of left phenotype mediated by Pitx2c in the right gonad is accompanied by an increase in the percentage of type C divisions that resembles that of wild-type left gonad (P = 0.0001; Figs. 1L, 2C, and 3 T and U and statistical analysis in Table S1).
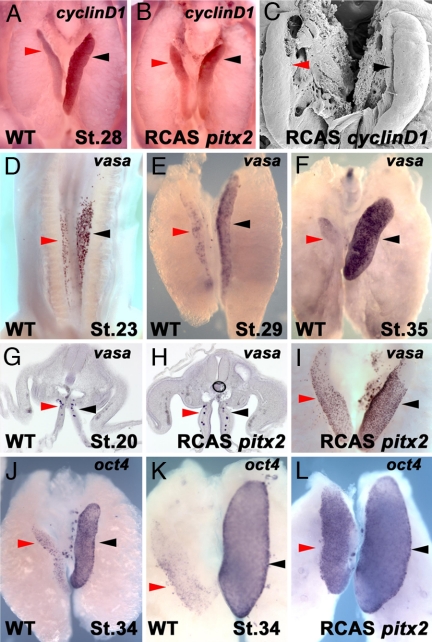
To ascertain whether the transcriptional activity induced by Pitx2c might affect the interactions between somatic and primordial germ cells (PGCs) we assayed the spatiotemporal transcript distribution of vasa, a characteristic functional marker of PGCs in vertebrates and invertebrates (31–36), oct4 (37), and nanog (38) transcripts. Avian PGCs originate in the epiblast and migrate into the germinal ridges via the bloodstream and through the gut and dorsal mesentery (39, 40). In situ hybridization at stage 20 HH to detect vasa transcripts in PGCs en route to the gonads do not show differences in either control embryos or in those with ectopic pitx2c expression (n = 52, 97%; Fig. 4 G and H). Differences in PGC number and pattern distribution start to be observed before overt morphological asymmetries between the left and right gonads (reviewed in ref. 41) at stages 22–23 HH (Fig. 4D) and are pronounced by stage 29 HH (Fig. 4E). Subsequently, whereas in the left gonad PGCs proliferate and increase in number, regression and size reduction of the somatic compartment of the right gonad correlate with a drastic diminution of vasa, oct4, and nanog transcripts (and thus PGC number) and eventual disappearance by stage 38 HH (Fig. 4 H and J and data not shown). Forced pitx2c expression in the right gonad is able to prevent the disappearance of PGCs because their number and pattern distribution mirror those of the left gonad (n = 65, 32%; Fig. 4 I and L). These results suggest that Pitx2 is not only involved in allowing normal somatic gonad cell growth and differentiation as described above, but is also able to regulate PGC proliferation and survival. Overall, our observations point out the existence of cellular interactions between the somatic and germ-line compartments required for correct gonad size and function and suggest a role for Pitx2c in mediating these interactions.
Fig. 4.
Pitx2c regulates PGC survival and proliferation. Asymmetric expression of cyclinD1 is detected at stage 28 HH (A). Expression of cyclinD1 is induced after pitx2c overexpression in the right gonad (B) when compared with a control (A/P-infected) right gonad in A. Overexpression of cyclinD1 induces right gonad outgrowth (C) when compared with a control (Fig. 3E). Histological sections showing symmetric vasa expression at the time when PGCs migrate toward the gonads in control embryos at stage 20 HH (G) and in pitx2c transfected gonads (H). Red and black arrowheads show symmetric PGC numbers in wild-type and pitx2 transfected embryos in G and H, respectively. (D and E) Asymmetric distribution of vasa transcripts at stage 23 HH (D) and stage 29 HH (E). vasa (F) and oct4 (J) transcripts are highly detected in the left gonad at stages 35 and 34 HH, respectively, but almost absent in the regressing right gonad at stage 38 (K and not shown for vasa). After pitx2c overexpression, vasa and oct4 expression is restored in the right gonad and detection of both transcripts is symmetrical (I and L, respectively). Red and black arrowheads points to the right and left gonad, respectively.
Pitx2 Controls Gonad Size Through cyclinD1 Activation.
Pitx2 has been shown to activate essential specific growth control gene targets acting in G1, including cyclinD2 (42–44). These observations prompted the search for growth-related genes that might be asymmetrically expressed during gonad development. Whereas cyclinD2 does not appear to display asymmetric expression (data not shown), our results indicate that transcripts for cyclinD1 are more abundant in the left than in the right developing cortex at stages 27/28 HH (Fig. 4A). These data suggest that cyclinD1 might be involved in the differential outgrowth observed between the female left and right gonads. This was tested by overexpressing cyclinD1 in the right urogenital ridge. As was the case for pitx2c, forced expression of cyclinD1 in the right gonad, even though it was not able to induce cortical differentiation (and thus was unable to completely rescue the degeneration of the right gonad), led to enhanced cell proliferation and gonad overgrowth (n = 55, 29%; Fig. 4C; compare with Fig. 3E). To test a possible interaction between CyclinD1 and Pitx2 we ectopically expressed pitx2c on the right urogenital ridge and analyzed cyclinD1 expression by in situ hybridization. Increased levels of cyclinD1 were mainly observed in the anterior right cortex (Fig. 4B). The converse experiment, overexpression of cyclinD1, did not induce pitx2c expression (data not shown). These results suggest that CyclinD1 is downstream of Pitx2. Similar results have recently been published (45).
These observations directly implicate Pitx2c and CyclinD1 as pivotal players in determining the final size, shape, and morphogenesis of the developing avian gonads. Combined with previous findings, in other cellular contexts where CyclinD1 is a direct target of Pitx2 (42, 43, 46, 47), our results suggest that the Pitx2–CyclinD pathway might be a prototypic module used to regulate cell-type-specific proliferation during embryogenesis. However, the fact that CyclinD1 is not sufficient to completely rescue the degeneration of the right gonad indicates the presence of other factors/mechanisms acting in parallel and/or in conjunction with CyclinD1, and downstream of Pitx2, during gonad outgrowth and differentiation.
Conclusions
The mechanisms underlying left–right organ determination have fascinated biologists for decades, but we still ignore the nature of events that link specific gene functions to particular development of processes during the establishment of the left–right axis. Integrating the differentiation of many different cells and cell types into specific spatial patterns along the left–right axis implies a series of events that include changes in cell adhesion and shape, the acquisition of cell fate, cell migration, cell proliferation, and cell differentiation. By focusing on the development of female chick gonads, we have provided evidence that a key determinant of organ left–right asymmetry in vertebrates, the transcription factor Pitx2, is capable of influencing gonad shape and size, and, consequently, organ function and morphogenesis, by controlling somatic cell morphology, extracellular matrix composition, spindle orientation, and cell proliferation. Furthermore, our results suggest that Pitx2c, in concert with CyclinD1, allows the somatic cell compartment to control primordial germ cell survival and proliferation and, as such, identify these two genes as part of the regulatory gene network that coordinates growth, size, and gonad function. A fertile and challenging area of research that may allow us to better understand in the near future how gene activity influences morphogenesis will be determining the upstream signals and downstream Pitx2 effectors that coordinate vertebrate gonad shape, size, and function.
Materials and Methods
Chicken Embryo Manipulation.
Chicken embryos (Rhode Island) were incubated at 37.5°C for 1.5–12 days. Embryos between 10 and 38 HH were fixed in 4% paraformaldehyde in PBS for 2 h and overnight for immunostainings and in situ hybridization, respectively. For scanning electron microscopy, embryos were fixed in 2.5% glutaraldehyde/cacodylate buffer (0.1 M, pH 7.4) for 2 h. All fixations were performed at 4°C. Manipulation of embryos for injection with RCAS constructs was performed at various time points. Eggs were windowed, and a slit was cut in the viteline membrane for embryo access. Viral suspensions were injected by using a borosilicate needle and a pressurized injector. After injections, eggs were sealed with transparent tape, incubated for different periods of time, and fixed as indicated above.
Scanning Electron Microscopy.
Chicken embryos were fixed in 2.5% glutaraldehyde/cacodylate buffer (0.1 M, pH 7.4) for 2 h at 4°C, washed three times in cacodylate buffer, and subsequently processed for critical point and sputtering following standard procedures. Sputtering of gonads was performed for 4 min and was repeated for an extra minute in a different position for stage 38 HH gonads. Imaging was performed in a JEOL JSM 6390LV scanning microscope.
Retroviral Constructs.
RCAS constructs were constructed as previously described in ref. 48 and references therein. pitx2c constructs were a gift from Y. Chen (Tulane University, New Orleans). Full-length CyclinD1 was obtained by PCR using the primers 5′-CTTATCGATAGAGCGAGCGAGAGACTGAC-3′ and 5′-TTATCGATTACTTCTCTTGCCAAGCAAACC-3′.
Efficiency of transfection in chicken embryonic fibroblasts was monitored by using 3 μg of RCAS construct plus 0.5 μg of pCAGGS-GFP and visualizing GFP production 24 h after treatment.
Probes and in Situ Hybridization.
The cyclinD1, nanog, and oct4 probes were obtained after PCR amplification of chicken RNA collected from embryos at stage 10, 21, and 28 HH. The vasa probe was obtained from chicken EST ChEST271n20.
The following primers were used for PCR amplification: cyclinD1, 5′-AGAGCGAGCGAGAGACTGAC-3′ and 5′-GTGCAGGAAAGGTCTGCTTC-3′; nanog, 5′-CTTCCAGCTCTGGGACACTC-3′ and 5′-GGTGCTCTGGAAGCTGTAGG-3′; oct4, 5′-GGGAATTCATGCATGTAAAAGCCAAA-3′ and 5′-GCTGCTGTTCAGTGGCTGCTGTTGTTC-3′.
PCR products were subsequently cloned into pGEMT-easy vector, and digoxigenin-labeled probes were synthesized according to standard protocols. In situ hybridization was performed as described (49), and alkaline phosphatase reactions were developed by using BM purple substrate from Roche. Embryos were imaged by using a Leica DFC490 digital camera attached to a Leica 716 Apo microscope.
Sexing of Chicken Embryos.
Embryos were sexed by amplification of the sex-linked CHD1 genes CHD-W and CHD-Z. For this, a small piece of embryo tissue was digested in alkaline buffer with proteinase K at 55°C overnight and genomic DNA was extracted. Genomic DNA was amplified with the primers 5′-GTTACTGATTCGTCTACGAGA-3′ and 5′-ATTGAAATGATCCAGTGCTTG-3′.
The PCR products were analyzed. For females, two bands at 450 and 600 bp were observed, and for males a unique band at 600 bp was detected.
Immunohistochemistry, BrdU Incorporation, and Toluidine Blue Stainings.
Immunohistochemistry for laminin (Chemicon), β-catenin (BD Biosciences), N-cadherin (Sigma), Phospho Histone H3 (Upstate), and α-tubulin (Sigma) were performed following the manufacturer's protocols for dilutions. Briefly, embryos were fixed in 4% paraformaldehyde for 2 h at 4°C and then maintained in PBS. Embryos were permeabilized with 0.1% Triton for 10 min after incubation with the primary antibody. Primary antibodies were incubated overnight, and secondary antibodies were incubated for 1 h at 37°C. Staining with DAPI (Invitrogen) for DNA and phalloidin for actin were carried out following standard procedures at a final dilution of 1.5 μg/ml and 16.5 nM, respectively.
BrdU incorporation was detected by using the Roche 5-Bromo-2′-deoxy-uridine Labeling and Detection Kit I. For BrdU incorporation into tissues, eggs were windowed and 100 μl of the BrdU solution (100 μg/μl) was pipetted directly on top of the embryos. Subsequently, eggs were sealed as previously described and incubated for 1 h at 37.5°C. Embryos were fixed in 4% paraformaldehyde for 2 h and then maintained in PBS. Detection of BrdU by a specific antibody conjugated to fluorescein was performed by incubating sections for 1 h at 37°C.
Toluidine blue staining was performed following standard methods. Three-micrometer semithin sections were treated for 2 min in Toluidine blue solution, and nuclei were differentiated in 1-Butanol.
Imaging of sections was performed with a Leica SP5A OBS confocal microscope using DPSS (diode-pumped solid-state lasers), argon, blue diode and HeNe lasers.
Supplementary Material
Acknowledgments.
We acknowledge the Nottingham and United Kingdom Chicken Consortium EST Project for providing us with vasa chicken EST. We thank Drs. S. Boue, M. Ibañes, Y. Kawakami, and A. Raya for their valuable discussions and insight; Dr. Y. Chen for pitx2c constructs; D. Mulero and A. Lázaro for their technical assistance; and M. Schwarz and K. Spitere for their help in preparing the manuscript. J.R.-L. was partially supported by a contract from the Fondo de Investigacion Sanitario (FIS) (Ref. CP05/00217), Instituto de Salud Carlos III, Ministerio de Educacion y Ciencia (MEC), and a grant from the same institution (Ref. PI061863). Work in the laboratory of J.C.I.B. was supported by grants from the G. Harold and Leila Y. Mathers Charitable Foundation, Fundacion Cellex, MEC (BFU2006–12247), and the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804904105/DCSupplemental.
References
- 1.Levin M, Johnson RL, Stern CD, Kuehn M, Tabin C. A molecular pathway determining left-right asymmetry in chick embryogenesis. Cell. 1995;82:803–814. doi: 10.1016/0092-8674(95)90477-8. [DOI] [PubMed] [Google Scholar]
- 2.Capdevila J, Vogan KJ, Tabin C, Izpisua Belmonte JC. Mechanisms of left-right determination in vertebrates. Cell. 2000;101:9–21. doi: 10.1016/S0092-8674(00)80619-4. [DOI] [PubMed] [Google Scholar]
- 3.Mercola M, Levin M. Left-right determination in vertebrates. Annu Rev Cell Dev Biol. 2001;17:779–805. doi: 10.1146/annurev.cellbio.17.1.779. [DOI] [PubMed] [Google Scholar]
- 4.Levin M. Left-right asymmetry in embryonic development: A comprehensive review. Mech Dev. 2005;122:3–25. doi: 10.1016/j.mod.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Raya A, Izpisua Belmonte JC. Left-right asymmetry in the vertebrate embryo: From early information to higher-level integration. Nat Rev Genet. 2006;7:283–293. doi: 10.1038/nrg1830. [DOI] [PubMed] [Google Scholar]
- 6.Shiratori H, Hamada H. The left-right axis in the mouse: From origin to morphology. Development. 2006;133:2095–2104. doi: 10.1242/dev.02384. [DOI] [PubMed] [Google Scholar]
- 7.Ai D, et al. Pitx2 regulated cardiac left-right asymmetry by patterning second cardiac lineage-derived myocardium. Dev Biol. 2006;296:437–449. doi: 10.1016/j.ydbio.2006.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campione M, et al. The homeobox gene Pitx2: Mediator of asymmetric left-right signaling in vertebrate heart and gut looping. Development. 1999;126:1225–1234. doi: 10.1242/dev.126.6.1225. [DOI] [PubMed] [Google Scholar]
- 9.Dagle JM, et al. Pitx2c attenuation results in cardiac defects and abnormalities of intestinal orientation in developing Xenopus laevis. Dev Biol. 2003;262:268–281. doi: 10.1016/s0012-1606(03)00389-0. [DOI] [PubMed] [Google Scholar]
- 10.Essner JJ, Branford WW, Zhang J, Yost HJ. Mesendoderm and left-right brain, heart and gut development are differentially regulated by pitx2 isoforms. Development. 2000;127:1081–1093. doi: 10.1242/dev.127.5.1081. [DOI] [PubMed] [Google Scholar]
- 11.Gage PJ, Suh H, Camper SA. Dosage requirement of Pitx2 for development of multiple organs. Development. 1999;126:4643–4651. doi: 10.1242/dev.126.20.4643. [DOI] [PubMed] [Google Scholar]
- 12.Gormley JP, Nascone-Yoder NM. Left and right contributions to the Xenopus heart: Implications for asymmetric morphogenesis. Dev Gene Evol. 2003;213:390–398. doi: 10.1007/s00427-003-0337-5. [DOI] [PubMed] [Google Scholar]
- 13.Kitamura K, et al. Mouse Pitx2 deficiency leads to anomalies of the ventral body wall, heart, extra-and periocular mesoderm and right pulmonary isomerism. Development. 1999;126:5749–5758. doi: 10.1242/dev.126.24.5749. [DOI] [PubMed] [Google Scholar]
- 14.Liu C, Liu W, Lu MF, Brown NA, Martin JF. Regulation of left-right asymmetry by thresholds of Pitx2c activity. Development. 2001;128:2039–2048. doi: 10.1242/dev.128.11.2039. [DOI] [PubMed] [Google Scholar]
- 15.Liu C, et al. Pitx2c patterns anterior myocardium and aortic arch vessels and is required for local cell movement into atrioventricular cushions. Development. 2002;129:5081–5091. doi: 10.1242/dev.129.21.5081. [DOI] [PubMed] [Google Scholar]
- 16.Logan M, Pagan-Westphal SM, Smith DM, Paganessi L, Tabin C. The transcription factor Pitx2 mediates situs-specific morphogenesis in response to left-right asymmetric signals. Cell. 1998;94:307–317. doi: 10.1016/s0092-8674(00)81474-9. [DOI] [PubMed] [Google Scholar]
- 17.Piedra ME, Icardo JM, Albajar M, Rodriguez-Rey JC, Ros MA. Pitx2 participates in the late phase of the pathway controlling left-right asymmetry. Cell. 1998;94:319–324. doi: 10.1016/s0092-8674(00)81475-0. [DOI] [PubMed] [Google Scholar]
- 18.Ryan AK, et al. Pitx2 determines left-right asymmetry of internal organs in vertebrates. Nature. 1998;394:545–551. doi: 10.1038/29004. [DOI] [PubMed] [Google Scholar]
- 19.Schweikert A, Campione M, Steinbeisser H. Blum M Pitx2 isoforms: involvement of Pitx2c but not Pitx2a or Pitx2b in vertebrate left-right asymmetry. Mech Dev. 90:41–51. doi: 10.1016/s0925-4773(99)00227-0. 2000. [DOI] [PubMed] [Google Scholar]
- 20.St Amand TR, et al. Cloning and expression pattern of chicken Pitx2: A new component in the SHH signaling pathway controlling embryonic heart looping. Biophys Res Commun. 1998;247:100–105. doi: 10.1006/bbrc.1998.8740. [DOI] [PubMed] [Google Scholar]
- 21.Yoshioka H, et al. Pitx2, a bicoid-type homeobox gene, is involved in a lefty-signaling pathway in determination of left-right asymmetry. Cell. 1998;94:299–305. doi: 10.1016/s0092-8674(00)81473-7. [DOI] [PubMed] [Google Scholar]
- 22.Kinsky FC. The consistent presence of paired ovaries in the Kiwi (Apteryx) with some discussion of this condition in other birds. J Ornithol. 1971;112:334–357. [Google Scholar]
- 23.Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo 1951. Dev Dyn. 1992;195:231–272. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- 24.Guioli S, Lovell-Badge R. PITX2 controls asymmetric gonadal development in both sexes of the chick and can rescue the degeneration of the right ovary. Development. 2007;134:4199–4208. doi: 10.1242/dev.010249. [DOI] [PubMed] [Google Scholar]
- 25.Sekido R, Lovell-Badge R. Mechanisms of gonadal morphogenesis are not conserved between chick and mouse. Dev Biol. 2007;302:132–142. doi: 10.1016/j.ydbio.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 26.Carlon N, Stahl A. Origin of the somatic components in chick embryonic gonads. Arch Anat Microsc Morphol Exp. 1985;74:52–59. [PubMed] [Google Scholar]
- 27.Lien WH, Klezovitch O, Vasioukhin V. Cadherin-catenin proteins in vertebrate development. Curr Opin Cell Biol. 2006;18:499–506. doi: 10.1016/j.ceb.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Lilien J, Balsamo J. The regulation of cadherin-mediated adhesion by tyrosine phosphorylation/dephosphorylation of beta-catenin. Curr Opin Cell Biol. 2005;17:459–465. doi: 10.1016/j.ceb.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 29.Baena-Lopez LA, Baonza A, Garcia-Bellido A. The orientation of cell divisions determines the shape of Drosophila organs. Curr Biol. 2005;15:1640–1644. doi: 10.1016/j.cub.2005.07.062. [DOI] [PubMed] [Google Scholar]
- 30.Gong Y, Mo C, Fraser SE. Planar cell polarity signalling controls cell division orientation during zebrafish gastrulation. Nature. 2004;430:689–693. doi: 10.1038/nature02796. [DOI] [PubMed] [Google Scholar]
- 31.Fujiwara Y, et al. Isolation of a DEAD-family protein gene that encodes a murine homolog of Drosophila vasa and its specific expression in germ cell lineage. Proc Natl Acad Sci USA. 1994;91:12258–12262. doi: 10.1073/pnas.91.25.12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Komiya T, Itoh K, Ikenishi K, Furusawa M. Isolation and characterization of a novel gene of the DEAD box protein family which is specifically expressed in germ cells of Xenopus laevis. Dev Biol. 1994;162:354–363. doi: 10.1006/dbio.1994.1093. [DOI] [PubMed] [Google Scholar]
- 33.Komiya T, Tanigawa Y. Cloning of a gene of the DEAD box protein family which is specifically expressed in germ cells in rats. Biochem Biophys Res Commun. 1995;207:405–410. doi: 10.1006/bbrc.1995.1202. [DOI] [PubMed] [Google Scholar]
- 34.Olsen LC, Aasland R, Fjose A. A vasa-like gene in zebrafish identifies putative primordial germ cells. Mech Dev. 1997;66:95–105. doi: 10.1016/s0925-4773(97)00099-3. [DOI] [PubMed] [Google Scholar]
- 35.Tsunekawa N, Naito M, Sakai Y, Nishida T, Noce T. Isolation of chicken vasa homolog gene and tracing the origin of primordial germ cells. Development. 2000;127:2741–2750. doi: 10.1242/dev.127.12.2741. [DOI] [PubMed] [Google Scholar]
- 36.Yoon C, Kawakami K, Hopkins N. Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell-stage embryos and is expressed in the primordial germ cells. Development. 1997;124:3157–3165. doi: 10.1242/dev.124.16.3157. [DOI] [PubMed] [Google Scholar]
- 37.Lavial F, et al. The Oct4 homologue PouV and Nanog regulate pluripotency in chicken embryonic stem cells. Development. 2007;134:3549–3563. doi: 10.1242/dev.006569. [DOI] [PubMed] [Google Scholar]
- 38.Canon S, Herranz C, Manzanares M. Germ cell restricted expression of chick Nanog. Dev Dyn. 2006;235:2889–2894. doi: 10.1002/dvdy.20927. [DOI] [PubMed] [Google Scholar]
- 39.Kuwana T, Rogulska T. Migratory mechanisms of chick primordial germ cells toward gonadal anlage. Cell Mol Biol (Noisy-le-grand) 1999;45:725–736. [PubMed] [Google Scholar]
- 40.Nakamura Y, et al. Migration and proliferation of primordial germ cells in the early chicken embryo. Poultry Sci. 20007;86:2182–2193. doi: 10.1093/ps/86.10.2182. [DOI] [PubMed] [Google Scholar]
- 41.Smith CA, Sinclair AH. Sex determination: Insights from the chicken. BioEssays. 2004;26:120–132. doi: 10.1002/bies.10400. [DOI] [PubMed] [Google Scholar]
- 42.Briata P, et al. The Wnt/beta-catenin→Pitx2 pathway controls the turnover of Pitx2 and other unstable mRNAs. Mol Cell. 2003;12:1201–1211. doi: 10.1016/s1097-2765(03)00407-6. [DOI] [PubMed] [Google Scholar]
- 43.Kioussi C, et al. Identification of a Wnt/Dvl/beta-Catenin→ Pitx2 pathway mediating cell-type-specific proliferation during development. Cell. 2002;111:673–685. doi: 10.1016/s0092-8674(02)01084-x. [DOI] [PubMed] [Google Scholar]
- 44.Wei Q, Adelstein RS. Pitx2a expression alters actin-myosin cytoskeleton and migration of HeLa cells through Rho GTPase signaling. Mol Biol Cell. 2002;13:683–697. doi: 10.1091/mbc.01-07-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishimaru Y, et al. Mechanism of asymmetric ovarian development in chick embryos. Development. 2008;135:677–685. doi: 10.1242/dev.012856. [DOI] [PubMed] [Google Scholar]
- 46.Baek SH, et al. Regulated subset of G1 growth-control genes in response to derepression by the Wnt pathway. Proc Natl Acad Sci USA. 2003;100:3245–3250. doi: 10.1073/pnas.0330217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martinez-Fernandez S, et al. Pitx2c overexpression promotes cell proliferation and arrests differentiation in myoblasts. Dev Dyn. 2006;235:2930–2939. doi: 10.1002/dvdy.20924. [DOI] [PubMed] [Google Scholar]
- 48.Raya A, et al. Notch activity acts as a sensor for extracellular calcium during vertebrate left-right determination. Nature. 2004;427:121–128. doi: 10.1038/nature02190. [DOI] [PubMed] [Google Scholar]
- 49.Rodriguez-Esteban C, et al. The novel Cer-like protein Carone mediates the establishment of embryonic left-right asymmetry. Nature. 1999;401:243–251. doi: 10.1038/45738. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.