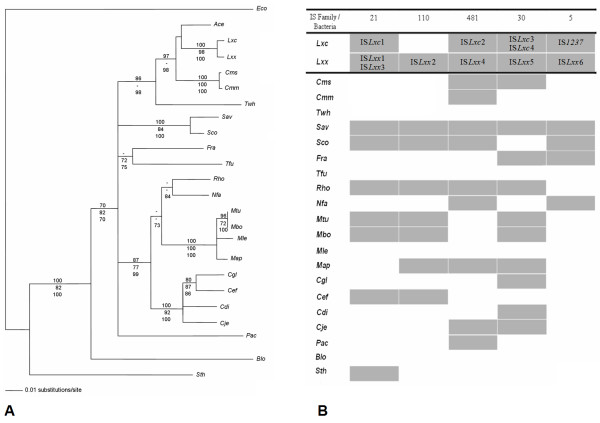
Figure 4.
Comparative analysis of Leifsonia xyli IS elements with putative transposases of Actinobacteria. (A) Actinobacteria were arranged based on their phylogenetic relatedness. Consensus neighbor-joining phylogenetic tree was constructed using SSU rDNA(16S) sequences of: twenty-two completely sequenced genomes; in addition to Agromyces sp., which was reported as forming a coherent cluster to Leifsonia [44,45]; and E. coli as a outgroup. Numbers at the branches represent bootstrap values for parsimony (top, 2000 replicates), maximum likelihood (middle, 100 replicates) and neighbor-joining (bottom, 2000 replicates). Only bootstrap values above 70 are shown. (B) IS elements characterized from Lxc and Lxx genomes were used as in silico probes for the identification of distribution of IS families belonging to Leifsonia xyli genomes in other Actinobacteria. The ISfinder database was also searched for the presence of IS families in the Actinobacteria assayed here. Species names and accession numbers are listed: Eco-Escherichia coli K12 (NC_000913); Ace – Agromyces sp. (AM410680); Lxc – Leifsonia xyli subsp. cynodontis DSMZ46306 (unpublished); Lxx – Leifsonia xyli subsp. xyli CTCB07 (NC_006087); Cms – Clavibacter michiganensis subsp. sepedonicus ATCC33113 (NC_010407); Cmm – Clavibacter michiganensis subsp. michiganensis NCPPB 382 (NC_009480); Twh – Tropheryma whipplei str. Twist (NC_004572) and TW08/27 (NC_004551); Sav – Streptomyces avermitilis MA-4680 (NC_003155); Sco – Streptomyces coelicolor A3(2) (NC_003888); Fra – Frankia sp. CcI3 (NC_007777); Tfu – Thermobifida fusca YX (NC_007333); Rho – Rhodococcus sp. RHA1 (NC_008268); Nfa – Nocardia farcinica IFM 10152 (NC_006361); Mtu – Mycobacterium tuberculosis H37Rv (NC_000962) and CDC1551 (NC_002755); Mbo – Mycobacterium bovis AF2122/97 (NC_002945); Mle – Mycobacterium leprae TN (NC_002677); Map – Mycobacterium avium subsp. paratuberculosis str. k10 (NC_002944); Cgl – Corynebacterium glutamicum ATCC 13032 (NC_006958); Cef – Corynebacterium efficiens YS-314 (NC_004369); Cdi – Corynebacterium diphtheriae NCTC 13129 (NC_002935); Cje – Corynebacterium jeikeium K411 (NC_007164); Pac – Propionibacterium acnes KPA171202 (NC_006085); Blo – Bifidobacterium longum NCC2705 (NC_004307); Sth – Symbiobacterium thermophilum IAM 14863 (NC_006177).