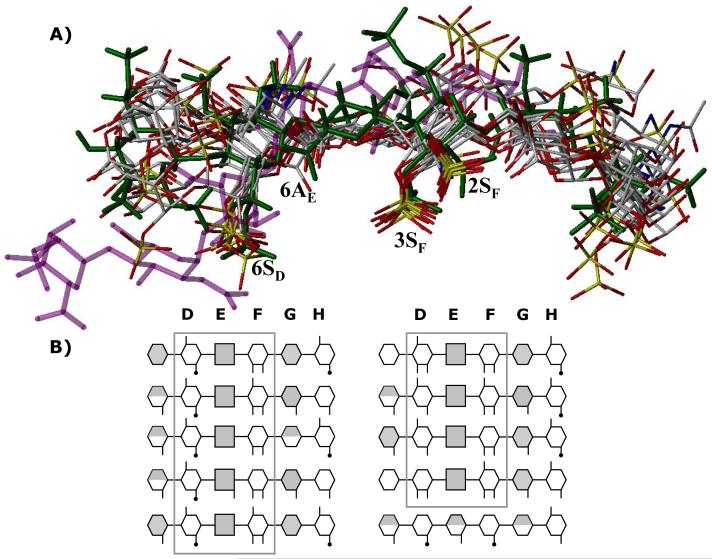
Figure 8. Finding a needle in a haystack.
A) An overlay of final 10 hexasaccharide sequences obtained after second phase of combinatorial library screening. Structure in green is the H5CRYS, while those in atom-type color are 9 sequences with nearly identical binding orientation and geometry. Sequence in purple color was found to bind antithrombin reproducibly with high specificity and affinity but dramatically different orientation. Labels 2SF, 3SF, 6AE and 6SD represent sulfate or carboxylate groups at the 2- and 3-position of residue F, 6-position of residue E and 6-positon of residue D. B) Symbolic representation of the high-affinity, high-specificity hexasaccharide structures shown above. The hexasaccharide library sequence runs {UAp(1→4)GlcNp(1→4)}3, where UA is either IdoAp (shaded hexagon) or GlcAp (shaded square). Sulfated substitution at 2-, 3- or 6-positions of either UAp or GlcNp is indicated with a line (—), while acetate substitution at the 2-positon of GlcNp is indicated with a line-dot (—·). Iduronic acid residues in 2SO conformation are shown as fully shaded hexagons, while those in 1C4 conformation are shown as half-filled hexagons. See text for details. See Supplementary Information for a MOL2 file of all the combinatorial sequences.