Figure 6.
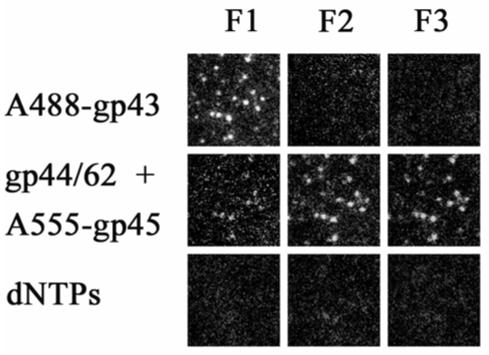
The binding of gp43 to forked DNA substrate, followed by gp45 and gp44/62, results in the formation of an active DNA polymerase holoenzyme. Each frame represents the fluorescence of single molecules of DNA with proteins bound in the order indicated at the side of each row. The filter sets are as described in the legend to Figure 2. The proteins were in 20 mM Tris, 5 mM magnesium acetate, 1 mM dithiothreitol, pH 7.9. For the addition of gp45 and gp44/62, 2.5 mM ATP was included. The loss of fluorescence upon the addition of dNTPs indicates the holoenzyme maintains strand displacement activity on single DNA molecules.
