Abstract
Viruses are detected by different classes of pattern recognition receptors (PRRs), such as Toll-like receptors and RIG-like helicases. Engagement of PRRs leads to activation of interferon (IFN)-regulatory factor 3 (IRF3) and IRF7 through IKKɛ and TBK1 and consequently IFN-β induction. Vaccinia virus (VACV) encodes proteins that manipulate host signalling, sometimes by targeting uncharacterised proteins. Here, we describe a novel VACV protein, K7, which can inhibit PRR-induced IFN-β induction by preventing TBK1/IKKɛ-mediated IRF activation. We identified DEAD box protein 3 (DDX3) as a host target of K7. Expression of DDX3 enhanced Ifnb promoter induction by TBK1/IKKɛ, whereas knockdown of DDX3 inhibited this, and virus- or dsRNA-induced IRF3 activation. Further, dominant-negative DDX3 inhibited virus-, dsRNA- and cytosolic DNA-stimulated Ccl5 promoter induction, which is also TBK1/IKKɛ dependent. Both K7 binding and enhancement of Ifnb induction mapped to the N-terminus of DDX3. Furthermore, virus infection induced an association between DDX3 and IKKɛ. Therefore, this study shows for the first time the involvement of a DEAD box helicase in TBK1/IKKɛ-mediated IRF activation and Ifnb promoter induction.
Keywords: DEAD box protein, immune evasion, interferon-β, IRF activation, vaccinia virus
Introduction
The innate immune response to viruses is critically dependent on their recognition by pattern recognition receptors (PRRs), which induce the production of type I interferons (IFNs). The different classes of PRRs implicated in this process include Toll-like receptors (TLRs), RIG-like helicases (RLHs) and cytosolic DNA receptors (Takaoka and Taniguchi, 2007). TLR3, 7, 8 and 9 constitute a subset of TLRs that localise to the endosomal compartment and recognise viral nucleic acids: TLR3 responds to dsRNA, TLR7 and 8 to ssRNA, and TLR9 to unmethylated CpG dsDNA (Bowie, 2007). In contrast, the RLHs recognise viral RNA in the cytoplasm. Retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (Mda-5) bind distinct viral dsRNAs and thereby recognise the different RNA viruses (Hornung et al, 2006; Kato et al, 2006; Pichlmair et al, 2006). An antiviral response can also be induced by cytoplasmic dsDNA through one or more receptor(s) that are independent of RIG-I and Mda-5 (Takaoka and Taniguchi, 2007). DNA-dependent activator of IFN-regulatory factors (IRFs) (DAI, also called DLM-1, ZBP1) has been implicated in this (Takaoka et al, 2007), although additional DNA receptors seem to be involved in vivo (Ishii et al, 2008).
All of these antiviral PRRs signal to the activation of transcription factors required for type I IFN induction, namely NF-κB, IRF3 and 7 (O'Neill, 2006). The upstream signalling pathways are distinct for different PRRs. Signalling from TLR3 and 4 to IRF3 and 7 activation involves the adaptor protein TRIF, whereas TLR7, 8 and 9 signal to these IRFs through MyD88 and interleukin-1 (IL-1) receptor-associated kinase 1 (IRAK1) (Moynagh, 2005). The RLHs signal to IRF activation through the CARD-domain-containing adaptor molecule MAVS (mitochondrial antiviral signalling, also called Cardif, VISA and IPS-1) (Yoneyama et al, 2004; Kawai et al, 2005; Meylan et al, 2005; Seth et al, 2005; Xu et al, 2005) while signalling adaptors for DAI and potential additional DNA receptor(s) have not yet been identified. Most of these PRR pathways (with the exception of TLR7, 8 and 9) converge at the level of IKKɛ and/or TBK1, the kinases that lead to the phosphorylation and activation of IRF3 and 7, placing them at the central point of the IRF activation pathways.
In the process of co-evolution with the host, viruses have developed sophisticated mechanisms to evade or subvert key aspects of the host antiviral response. Therefore, the study of viral evasion mechanisms can help to identify novel aspects of the host antiviral response. For example, the description of the vaccinia virus (VACV) TLR antagonists A46 and A52 provided evidence for the involvement of TLRs in the antiviral response (Bowie et al, 2000; Harte et al, 2003; Stack et al, 2005). A52 interacts with the TLR signalling components TNF-receptor-associated factor 6 (TRAF6) and IRAK2 (Harte et al, 2003; Maloney et al, 2005). By interacting with TRAF6, A52 can stimulate p38 MAP kinase activation and enhance TLR-mediated induction of the anti-inflammatory cytokine IL-10 (Maloney et al, 2005), whereas it inhibits TLR-induced NF-κB activation and pro-inflammatory cytokine production through targeting of IRAK2 (Keating et al, 2007).
We became interested in another as yet uncharacterised VACV protein, K7, because it shares significant sequence similarity with A52 and is very well conserved within the poxvirus family. Here, we show that K7 mimics some of the effects of A52 and shares its intracellular targets. However, unlike A52, K7 also has a potent inhibitory effect on TLR-dependent and -independent IRF activation and consequently Ifnb promoter induction, which suggests it might target additional host proteins. We identify the DEAD box protein (DDX3) as a target of K7 and show that DDX3 has a novel role in Ifnb promoter induction. DDX3 is an RNA helicase, and the human protein and/or its orthologues have previously been implicated in diverse cellular processes, including splicing, translational regulation, nuclear export of RNA (Rosner and Rinkevich, 2007) and more recently in cancer biogenesis (Botlagunta et al, 2008). Interestingly, DDX3 is also targeted by other viral proteins, namely hepatitis C virus (HCV) core protein (Mamiya and Worman, 1999; Owsianka and Patel, 1999; You et al, 1999) and HIV Rev (Yedavalli et al, 2004). HIV exploits the nucleocytoplasmic shuttling capacity of DDX3 for exporting its mRNAs out of the nucleus (Yedavalli et al, 2004), and DDX3 may also be required for HCV replication (Ariumi et al, 2007).
In this study, expression of DDX3 led to enhanced TBK1/IKKɛ-dependent Ifnb promoter induction, whereas suppression of DDX3 expression by RNAi inhibited this, as well as virus or poly(I:C)-induced IRF3 activation. This function of DDX3 required its N-terminal region, which correlated with the region that was targeted by K7. Further, virus infection stimulated the association of DDX3 and IKKɛ. Thus, we demonstrate an unexpected novel role for the DEAD box helicase DDX3 in Ifnb promoter induction and provide evidence that this function is targeted by a viral evasion strategy.
Results
Identification of K7
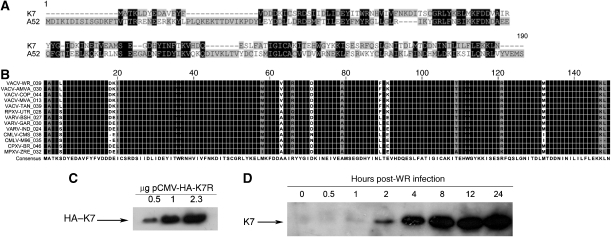
K7 was identified from an analysis of poxvirus genomes as having significant sequence similarity to the TLR antagonist A52. A52 (VACV_WR178) is a 190 amino-acid (aa) protein, whereas K7 (VACV_WR039) is only 149 aa long. An alignment of A52 and K7 from the VACV strain Western Reserve (WR) shows 26% aa identity and 45% similarity within the shared region of sequence (Figure 1A). Analysis of available K7 orthopoxvirus sequences (www.poxvirus.org) showed that K7 is highly conserved between different viruses, displaying greater than 95% aa identity between orthologues (Figure 1B), suggesting an important function for K7.
Figure 1.
Identification and expression of K7. (A) Alignment of A52 (VACV_WR178) and K7 (VACV_WR039) proteins from VACV (WR strain). (B) Alignment of K7 orthologues. VACV-WR: VACV Western Reserve; VACV-AMVA: VACV-Acambis 3000 MVA; VACV-COP: VACV Copenhagen; VACV-MVA: VACV modified virus Ankara; VACV-TAN: VACV Tian Tan; RPXV-UTR: rabbitpox virus Utrecht; VARV-BSH: variola virus Bangladesh; VARV-GAR: variola virus Garcia; VARV-India, variola virus India; CMLV-CMS: camelpox virus CMS; CMLV-M96: camelpox virus M96; CPXV-BR: cowpox virus Brighton red; MPXV-ZRE: monkeypox virus Zaire. (C) VACV WR K7R was cloned into pCMV-HA. Increasing amounts of pCMV-HA-K7R DNA were transfected into HEK293T cells, and 48 h later K7 expression was analysed by immunoblotting using an anti-HA antibody. (D) HEK293 cells infected with VACV WR at 10 p.f.u. per cell were harvested at the indicated times post-infection and lysates were analysed by immunoblotting using K7-specific antiserum.
To characterise the function of K7, K7R from VACV strain WR (VACV_WR039) was cloned into a mammalian expression vector containing an N-terminal HA epitope (pCMV-HA-K7R). Transfection of increasing amounts of pCMV-HA-K7R into HEK293T cells led to a concomitant increase in K7 expression at its predicted molecular mass of 17.5 kDa (Figure 1C). We then generated a rabbit polyclonal antiserum against K7 to confirm K7 protein expression in VACV WR-infected HEK293T cells. The expression of K7 was visible as early as 2 h post-infection (p.i.) and increased until 24 h p.i. (Figure 1D).
K7 inhibits TLR-induced NF-B activation and gene expression
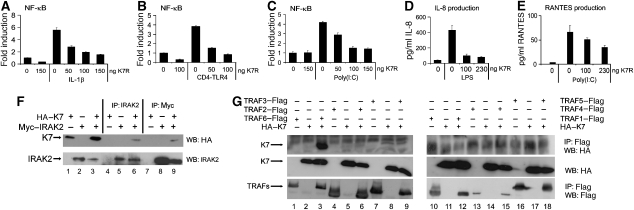
Given its similarity to A52, we tested whether K7 affected NF-κB activation induced by IL-1R/TLR signalling. Similar to A52, K7 expression inhibited IL-1-induced activation of the NF-κB reporter in a dose-dependent manner (Figure 2A). Also similar to A52, expression of K7 inhibited NF-κB activation induced by TLR4 (the constitutively active CD4-TLR4 fusion receptor) (Figure 2B) and TLR3 (activated by its synthetic dsRNA ligand poly(I:C) (Figure 2C). K7 also inhibited NF-κB-dependent chemokine expression, namely LPS-induced IL-8 production (Figure 2D) and poly(I:C)-induced RANTES production (Figure 2E).
Figure 2.
K7 inhibits TLR-induced NF-κB activation. In (A–C), HEK293 cells were transfected with pRK5-K7R or empty vector and the NF-κB luciferase reporter gene. Cells were stimulated with 20 ng/ml IL-1β for 6 h (A), transfected with 50 ng of CD4TLR4 for 24 h (B) or HEK293-TLR3 cells were stimulated with 25 μg/ml poly(I:C) for 8 h (C). Data are expressed as the mean fold induction±s.d. relative to control levels, for a representative experiment of three, each performed in triplicate. (D, E) For agonist-induced cytokines, HEK-TLR4 (D) or HEK-TLR3 (E) cells were transfected with K7R 24 h prior to stimulation with 1 μg/ml LPS or 25 μg/ml poly(I:C), respectively. After 24 h, supernatants were assayed for IL-8 (D) or RANTES (E) by ELISA. The experiments were performed four times in triplicate and data are expressed as the mean±s.d. from one representative experiment. (F) HEK293T cells were transfected with pCMV-HA-K7R and IRAK2-Myc and 48 h later, lysates were subjected to immunoprecipitation (IP) analysis with the indicated antibodies. (G) HEK293T cells were transfected with pCMV-HA-K7R and Flag–TRAF1, 2, 3, 4, 5 or 6 and 48 h later, lysates were subjected to IP analysis with the indicated antibodies. Results shown are representative of at least three experiments.
As both K7 and A52 affected the TLR–NF-κB axis, we investigated whether K7 shares intracellular targets with A52, namely IRAK2 and TRAF6 (Harte et al, 2003). K7 co-immunoprecipitated with IRAK2 when both proteins were co-expressed in HEK293T cells (Figure 2F, upper panel, lanes 6 and 9). Similarly, K7 co-immunoprecipitated with TRAF6 (Figure 2G, upper panel, lane 3), but did not interact with TRAFs 1–5 (Figure 2G, upper panel, lanes 6, 9, 12, 15 and 18). Similar to A52 (Harte et al, 2003), K7 co-immunoprecipitated with the TRAF domain of TRAF6 (aa 289–522) (not shown).
K7, but not A52, inhibits IRF activation induced by TLR and non-TLR pathways
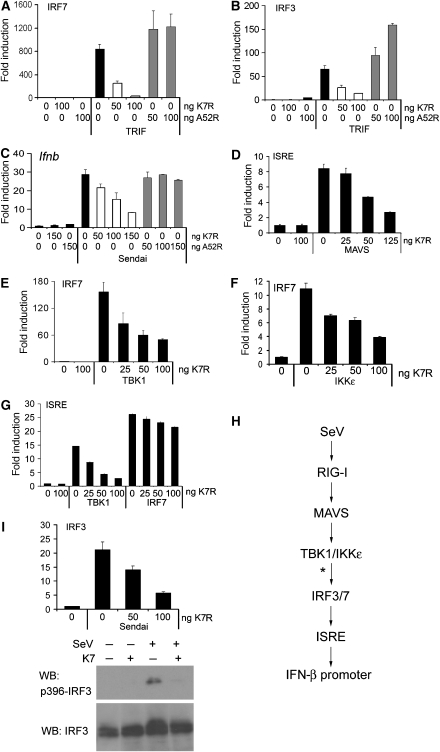
In contrast to the seemingly redundant effect of K7 and A52 on the TLR–NF-κB axis, a crucial difference between the two proteins became apparent when we investigated the effect of K7 on the signalling pathway leading to IRF activation. TLR3 and 4 stimulate IRF3 and 7 activation through TRIF, which is insensitive to A52 inhibition (Figure 3A and B), as TRAF6 and IRAK2 are not involved in this pathway (Keating et al, 2007). K7 strongly inhibited TRIF-induced activation of IRF3 and 7 (Figure 3A and B), thus revealing a key difference between A52 and K7.
Figure 3.
K7 inhibits IRF activation at the level of TBK1/IKKɛ. In (A–G), HEK293 cells were transfected with pRK5-K7R or pRK5-A52R, together with the indicated luciferase reporter genes for 24 h. Data are expressed as the mean fold induction±s.d. relative to control levels, for a representative experiment of at least two, each performed in triplicate. (A, B) Cells were transfected with IRF7-GAL4 (A) or IRF3-GAL4 (B) expression plasmids, and the GAL4-dependent pFR luciferase reporter gene plus 50 ng expression plasmid encoding TRIF. (C) Cells were transfected with the Ifnb promoter reporter gene prior to sendai virus infection for 16 h. (D) Cells were transfected with the ISRE reporter gene plus 50 ng MAVS expression plasmid. (E, F) Cells were transfected with IRF7-GAL4 and the pFR luciferase reporter gene, plus 50 ng of either TBK1 (E) or IKKɛ (F) expression plasmid. (G) Cells were transfected with the ISRE reporter gene plus 50 ng of either TBK1 or IRF7 expression construct for 24 h. (H) Schematic of SeV signalling pathway to Ifnb promoter induction. The deduced point of inhibition by K7 is marked with an asterisk. (I) For the IRF transactivation assay, cells were transfected as in (B), followed by stimulation with SeV for 16 h (upper panel). For immunoblot analysis, cells were transfected with K7, followed by SeV stimulation for 6 h. Western blots were probed for total IRF3 and phosphorylated IRF3 (Ser396) (lower panel).
We next tested whether K7 also inhibits non-TLR pathways to IRF3 and 7 activation. Upon recognition of its RNA through RIG-I, sendai virus (SeV) activates IRF3 and 7, and consequently induces the Ifnb promoter (Kato et al, 2005). Infection of cells with SeV led to induction of the Ifnb promoter, which was strongly inhibited by K7, but not A52 (Figure 3C). To define the site of action of K7 on this pathway, we examined the effect of K7 on each of the steps outlined in Figure 3H. To this end, we activated a reporter gene under the control of the IFN-stimulatory response element (ISRE) by expressing the MAVS adaptor protein that links RIG-I to IRF3/7. This was inhibited by K7 (Figure 3D). Downstream of MAVS, TBK1 and IKKɛ directly phosphorylate IRF3 and 7, which leads to their homo-dimerisation and activation (McWhirter et al, 2004). K7 inhibited TBK1-induced activation of IRF3 (not shown) and IRF7 (Figure 3E), and IKKɛ-induced IRF7 activation (Figure 3F), suggesting that K7 exerts an effect on TBK1/IKKɛ or further downstream. Consistent with this, K7 inhibited TBK1-induced ISRE activation, whereas direct induction of the ISRE by IRF7 expression was unaffected (Figure 3G). This pinpoints the inhibitory effect of K7 to the events around the activation of IRF3/7 by TBK1/IKKɛ (as shown by an asterisk in Figure 3H). K7 also inhibited SeV-stimulated IRF3 transactivation (Figure 3I). Further, SeV induced the appearance of higher molecular weight bands of endogenous IRF3, which represent phosphorylated forms. This was inhibited by K7, as was the appearance of virus-induced phospho-Ser396 IRF3 (Figure 3I). Thus, K7 can inhibit phosphorylation of endogenous IRF3.
K7 targets the host DEAD box protein DDX3
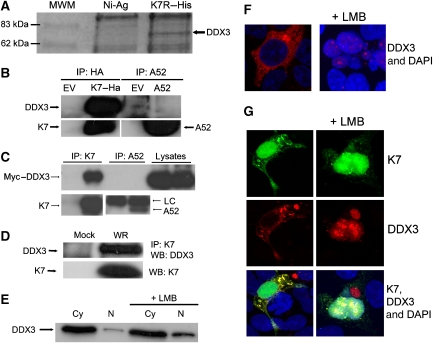
The inhibitory effects of K7 on IRF activation cannot be explained by its interaction with IRAK2 and TRAF6. We also failed to show an interaction between K7 and IKKɛ and/or TBK1 in co-immunoprecipitation experiments (not shown). Therefore, we reasoned that K7 must have additional host target(s) that can explain its ability to inhibit TBK1/IKKɛ-mediated IRF3/7 activation. To identify these, we used recombinant His-tagged K7 protein to isolate interacting proteins from HEK293 cell lysates and analysed these by SDS–PAGE. In a one-dimensional Coomassie-stained gel, we detected a band of approximately 70 kDa that appeared reproducibly in the K7 pull-down lane but not in the control lane (Figure 4A). This band was identified by peptide mass fingerprinting as DDX3 (also called DBX or Cap-Rf), a DEAD box-containing putative RNA helicase of 73 kDa. Human DDX3 and/or its orthologues have been implicated in several different cellular processes, including control of cell growth, translational control and nuclear–cytoplasmic shuttling (see Introduction).
Figure 4.
K7 but not A52 interacts with DDX3. (A) HEK293 cell lysates were added to purified His–K7 coupled to Ni-Agarose and incubated for 2 h at 4°C. The immune complexes were precipitated, subjected to SDS–PAGE and stained with Coomassie blue. A band of approximately 70 kDa (marked with an arrow) that appeared specifically in the K7 pull-down lane was excised, prepared for MALDI-TOF analysis and shown to be DDX3. (B) HEK293T cells were transfected with HA–K7R, A52R or empty vector (EV) and 48 h later, lysates were subjected to immunoprecipitation analysis to detect endogenous DDX3. (C) HEK293 cells were transfected with Myc–DDX3 expression plasmid. After 24 h cells were mock infected or infected with WR at 10 p.f.u. per cell, and 8 h later lysates were subjected to immunoprecipitation analysis with K7- or A52-specific antiserum LC: antibody light chain. (D) HEK293 cells were mock infected or infected with WR at 10 p.f.u. per cell and 8 h later, cell lysates were subjected to immunoprecipitation with K7-specific antiserum, followed by immunoblotting with the indicated antibodies. (E) HEK293 cells were either left untreated or incubated with 25 mM leptomycin B (LMB) for 4 h. Cytoplasmic (Cy) and nuclear (N) extracts were analysed for DDX3 by immunoblotting. (F) HEK293 cells were grown on 22-mm coverslips in six-well plates and transfected with the HA–DDX3 expression plasmid for 48 h and then 25 mM LMB was added. Cells were fixed 4 h later, permeabilised and stained with anti-HA-AlexaFluor594 and the DAPI nuclear stain. The slides were examined by phase-contrast and confocal microscopy. A section of approximately 1 μm through the centre of a cell is shown. (G) HEK293 cells were grown as in (F) and transfected with HA–DDX3 and K7–EYFP expression plasmids for 48 h. LMB (25 mM) was then added and the cells were fixed 4 h later, permeabilised and stained with anti-HA-AlexaFluor594 and DAPI. The slides were examined by phase contrast and confocal microscopy. A section of approximately 1 μm through the centre of cells is shown. Results shown for (A–G) are representative of at least two experiments.
To confirm the K7–DDX3 interaction by an alternative method and to test whether DDX3 was a K7-specific target, HA-tagged K7 or A52 was transfected into HEK293T cells and co-immunoprecipitation experiments were performed. Endogenous DDX3 co-precipitated with HA–K7 (Figure 4B, upper panel, lane 2), but not with A52 (Figure 4B, upper panel, lane 4). Also, in VACV-infected cells, Myc-tagged DDX3 co-immunoprecipitated with virally expressed K7 but not A52 (Figure 4C), confirming that DDX3 is a target for K7 but not A52. Figure 4D shows that endogenous DDX3 co-immunoprecipitated with K7 expressed during VACV infection, confirming that this interaction occurs with physiological levels of both proteins.
K7 can interact with DDX3 in the cytoplasm and nucleus
It has been reported that DDX3 shuttles between the cytoplasm and the nucleus, and that it is exported from the nucleus through the CRM-1 system (Yedavalli et al, 2004). We confirmed this property of DDX3 by immunoblot analysis of subcellular fractions: in the absence of the CRM-1 inhibitor leptomycin B (LMB) DDX3 was detected mostly in the cytoplasmic fraction, whereas treatment with LMB led to its accumulation in the nucleus (Figure 4E). This result was also confirmed by immunofluorescent staining of HA-tagged DDX3. In the absence of LMB, DDX3 was located in the cytoplasm and cannot be detected in the nucleus with this method. However, 4 h after the addition of LMB, DDX3 was largely only detected in the nucleus (Figure 4F).
We then wanted to determine whether K7 interacts with DDX3 in the cytoplasm, the nucleus or both. To this end, we cloned K7 into an expression vector that generates a K7–EYFP (enhanced yellow fluorescent protein) fusion protein. When we analysed the cellular localisation of K7–EYFP (Figure 4G) or HA-tagged K7 that was stained with a fluorescently labelled antibody against the HA tag (not shown), K7 was present in both the cytoplasm and the nucleus. In the absence of LMB, K7 and DDX3 colocalised in distinct regions of the cytoplasm (Figure 4G, left column). After CRM-1 inhibition by LMB, DDX3 accumulated in the nucleus where it again colocalised with K7 (Figure 4G, right column). As a control for our experimental set-up, we co-expressed A52–EYFP and DDX3 and found no colocalisation of A52 and DDX3 (not shown). Our data suggest that K7 and DDX3 can interact in the cytoplasm. The observed colocalisation in the nucleus could be real or due to the high levels of both proteins in the nucleus after LMB treatment.
DDX3 is a positive effector of the IRF pathway at the level of TBK1/IKK
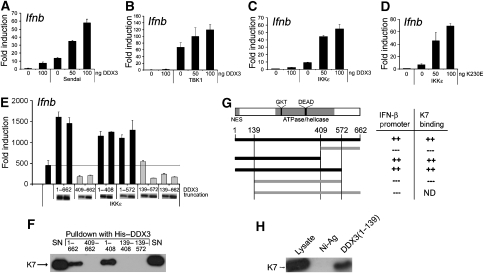
On the basis of our finding that K7 and not A52 can inhibit IRF activation and the identification of DDX3 as a K7-specific target, we wondered whether DDX3 itself has a role in this pathway. Ectopic expression of DDX3 alone led to some Ifnb promoter induction, whereas it strongly enhanced Ifnb promoter induction in response to SeV infection (Figure 5A), as opposed to the inhibitory effect observed for K7 expression (Figure 3C). Similar to the inhibitory effect of K7, the positive effect of DDX3 on Ifnb promoter induction localised to the level of TBK1/IKKɛ, as DDX3 expression also enhanced TBK1- and IKKɛ-mediated Ifnb promoter induction (Figure 5B and C). Given that DDX3 is a helicase, we wondered whether this activity was important for its ability to stimulate the Ifnb promoter. To address this, we generated a DDX3 point mutant by changing lysine at position 230 in a critical ATPase motif to glutamic acid. This K230E mutant has previously been shown to lose the ability to hydrolyse ATP and unwind RNA (Yedavalli et al, 2004). The expression of K230E enhanced activation of the Ifnb promoter by IKKɛ as potently as wild-type DDX3 (compare Figure 5C and D), suggesting that the effect of DDX3 on Ifnb promoter induction is independent of its ATPase/helicase activity.
Figure 5.
Role for the N terminus of DDX3 in TBK1/IKKɛ-induced Ifnb promoter activation. In (A–C), HEK293 cells were transfected with the indicated amount of pCMV-Myc-DDX3 (nanograms) together with the Ifnb promoter reporter gene, cells were harvested after 24 h and luciferase reporter gene activity was measured. (A) Cells were infected with sendai virus for 16 h. (B) Cells were transfected with 50 ng TBK1 expression plasmid. (C) Cells were transfected with 50 ng IKKɛ expression plasmid. (D) Cells were transfected with the Ifnb promoter reporter gene, together with 50 ng IKKɛ and the indicated amount (nanograms) of an expression plasmid encoding a mutant version of DDX3 lacking ATPase activity (K230E). (E) Cells were transfected with the Ifnb promoter reporter gene, together with 50 ng IKKɛ and 50 or 100 ng of expression vectors encoding full-length DDX3 (1–662) or the indicated N- and/or C-terminally truncated DDX3 proteins. Expression of DDX3 truncations by immunoblot is also shown. For (A–E), data are expressed as the mean fold induction±s.d. relative to control levels, for a representative experiment of at least two, each performed in triplicate. (F) Either full-length recombinant His-tagged DDX3 (1–662) or different His-tagged truncated DDX3 proteins were used in pull-down assays with cell lysates from HEK293T cells transfected with the HA–K7 expression plasmid. The precipitates were then analysed by immunoblotting with anti-HA antibody. Results are representative of three experiments. (G) Schematic of DDX3 showing in grey the positions of the nuclear export sequence (NES) and the conserved ATPase/helicase domain. The DEAD motif and the GKT motif that was mutated in the K230E point mutant are also indicated. A schematic summarising the different truncation mutants of DDX3 and their abilities to stimulate Ifnb promoter activation and to bind to K7, both of which depend on the presence of the N-terminus of DDX3 (aa 1–139), is shown below. (H) The N-terminus of DDX3 (aa 1–139) was expressed as a His-tagged recombinant protein and used in pull-down assays with cell lysates from HEK293T cells transfected with HA–K7. The precipitates were analysed by immunoblotting with anti-HA antibody. Result is representative of two experiments.
The N-terminus of DDX3 is required for Ifnb promoter induction and confers K7 binding
DEAD box helicases display a relatively high degree of sequence similarity across the two central RecA-like domains that contain the conserved helicase motifs. However, the N- and C-terminal flanking regions are more divergent and thought to confer specificity of function (Hogbom et al, 2007). As the ATPase and helicase activity of DDX3 seemed to be dispensable for its effect on the Ifnb promoter (Figure 5D), we wondered whether the N- and C-terminal regions mediate this novel function of DDX3. Hence, we generated DDX3 truncation mutants that were either lacking the C-terminal (1–572), the N-terminal (139–662) flanking region or both (139–572). Further truncations were made that divide the protein between the two RecA domains: 1–408 and 409–662.
When we analysed these mutants for their effects on IKKɛ-induced Ifnb promoter induction, we found that the C-terminal truncations (1–572, 1–408) enhanced promoter induction similar to the full-length DDX3 (1–662) (Figure 5E, black bars). However, all the mutants truncated at the N-terminus (409–662, 139–662 and 139–572) failed to enhance Ifnb promoter induction and even had inhibitory effects (Figure 5E, grey bars). These data show that the N-terminal region between aa 1–139 is required for the positive effect of DDX3 on Ifnb promoter induction.
Interestingly, we found that this region of DDX3 also confers binding to K7. This was demonstrated using recombinant His-tagged truncation mutants of DDX3 that correspond to the expression plasmids used for studying the effects on the Ifnb promoter. With His-tagged full-length DDX3 and the truncated DDX3 versions, we performed pull-down assays with cell lysates from HEK293T cells that had been transfected with HA–K7. Full-length DDX3 as well as DDX3(1–408) interacted with K7, whereas the DDX3 truncations lacking the N terminus (409–662, 139–572 and 139–408) failed to bind K7 (Figure 5F). These data strongly suggest that K7 binds to the N-terminal region of DDX3 (aa 1–139) that is required for the enhancement of the Ifnb promoter. This correlation is summarised in Figure 5G.
To further confirm that K7 targets the N-terminal region of DDX3, we generated recombinant His-tagged DDX3 comprising only aa 1–139 and repeated the pull-down experiment. As shown in Figure 5H, the N-terminus of DDX3(1–139) was sufficient for K7 binding.
The N-terminus of DDX3 exerts an effect as a dominant negative to suppress IRF-dependent promoter induction
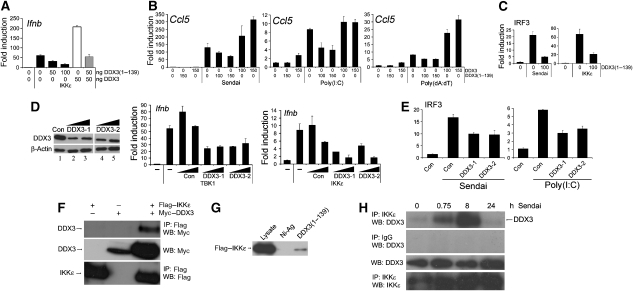
As the N-terminal region of DDX3 seemed to be crucial for its positive effect on the Ifnb promoter, we wondered whether the expression of DDX3(1–139) would have a dominant-negative effect. Indeed, co-expression of DDX3(1–139) inhibited IKKɛ-induced activation of the Ifnb promoter (Figure 6A, black bars) and reversed the enhancement observed with the expression of full-length DDX3 (Figure 6A, white and grey bars). This suggested that the N-terminus of DDX3 might be able to inhibit DDX3 function in other TBK1/IKKɛ-dependent pathways that involve endogenous DDX3. Hence, we examined the effect of DDX3(1–139) on different PRR pathways leading to the induction of the IRF3-dependent promoter Ccl5 (RANTES). Figure 6B shows that SeV (through RIG-I), poly(I:C) (through mda-5) and poly (dA:dT) (through the cytoplasmic DNA receptor(s)), all induced the Ccl5 promoter. In each case, this was inhibited by the expression of DDX3(1–139). In contrast, full-length DDX3 enhanced promoter induction (Figure 6B). SeV- or IKKɛ-stimulated IRF3 activation was also inhibited by DDX3(1–139) (Figure 6C), confirming an effect on IRF activation. These data suggest a requirement for DDX3 for full induction of IRF-dependent promoters activated by PRR signalling.
Figure 6.
DDX3 is required for TBK1/IKKɛ-dependent promoters and is recruited to IKKɛ. (A) HEK293 cells were transfected with the indicated amount (nanograms) of pCMV-DDX3 and/or pCMV-DDX3(1–139), 50 ng IKKɛ expression plasmid and the Ifnb promoter reporter gene. (B) Cells were transfected with a Ccl5 (RANTES) promoter reporter gene and the indicated amount (nanograms) of either pCMV-DDX3 or pCMV-DDX3(1–139) 24 h prior to stimulation with sendai virus or transfection with either poly(I:C) (5 μg per well) or poly(dA:dT) (1 μg per well). Cells were harvested 16 h later. (C) Cells were transfected with 0 or 100 ng pCMV-DDX3(1–139) together with the IRF3 reporter genes, and either infected with sendai virus (left panel) or transfected with IKKɛ (right panel). (D) DDX3 expression was suppressed using two different RNAi oligonucleotides, DDX3-1 and DDX3-2. HEK293 cells were transfected on two consecutive days with the DDX3 RNAi oligonucleotides or a control oligonucleotide matched for GC content. At 48 h after the second transfection, the cell lysates were analysed by immunoblotting with the indicated antibodies to confirm the reduction of endogenous DDX3 protein levels (left panel). HEK293 cells were co-transfected with the Ifnb promoter reporter gene and 50 ng expression plasmid for either TBK1 (middle panel) or IKKɛ (right panel) during the second transfection with RNAi oligonucleotides. (E) Similar to (D), except cells were transfected with IRF3 reporter genes and stimulated with sendai virus (left panel) or poly(I:C) (right panel). In (A–E), reporter gene activity was measured and data are expressed as the mean fold induction±s.d. relative to control levels, for a representative experiment of at least three, each performed in triplicate. (F) HEK293T cells were transfected with Myc–DDX3 and Flag–IKKɛ for 48 h and the lysates were analysed by immunoprecipitation (IP) with the indicated antibodies. (G) Recombinant His–DDX3(1–139) was used in ‘pull-down' assays with lysates from HEK293T cells transfected with Flag–IKKɛ. The precipitates were analysed by immunoblotting with Flag Ab. (H) HEK293T cells were infected with sendai virus and samples were harvested at the indicated times after virus infection. The cell lysates were prepared, immunoprecipitated with anti-IKKɛ antibody and analysed by immunoblotting with the indicated antibodies. All results are representative of two or more experiments.
RNAi depletion of DDX3 inhibits TBK1- and IKK-mediated Ifnb promoter induction
To confirm the role of endogenous DDX3 in TBK1/IKKɛ-mediated IRF-dependent promoter induction, we suppressed endogenous DDX3 expression using RNAi. Two oligonucleotides specifically targeting Ddx3 led to a significant reduction in DDX3 protein expression when compared to cells transfected with a control oligonucleotide (Figure 6D). Similar to previous investigators (Yedavalli et al, 2004), we found it difficult to achieve complete knockdown of DDX3 protein expression, which may be due to the high levels of DDX3 in the cell and/or the stability of the protein. Nonetheless, we observed reduced TBK1- and IKKɛ-induced activation of the Ifnb promoter when endogenous DDX3 expression was reduced by either RNAi oligonucleotide (Figure 6D). Furthermore, DDX3 RNAi inhibited SeV- or poly(I:C)-stimulated IRF3 activation (Figure 6E). This provides further evidence for the involvement of endogenous DDX3 in TBK1/IKKɛ-mediated activation of IRF3.
DDX3 interacts with IKK in a virus-dependent manner
Our data so far consistently pointed to the involvement of DDX3 in the activation of IRF3/7 by TBK1/IKKɛ, but it remained to be determined whether DDX3 was actually part of this signalling complex. To address this, the interaction between Flag–IKKɛ and Myc–DDX3 was tested in a co-immunoprecipitation assay, where Myc–DDX3 co-immunoprecipitated with Flag–IKKɛ (Figure 6F). Interestingly, in the presence of IKKɛ, we also observed a higher molecular weight form of DDX3 in the cell lysates (Figure 6F, middle panel), which might represent a phosphorylated form of DDX3.
As it is the N-terminal region of DDX3 that mediates the positive effect on the Ifnb promoter, we wondered whether IKKɛ binds to this region of DDX3. In a pull-down assay with recombinant His–DDX3(1–139), we found that IKKɛ can indeed bind to the isolated N-terminus of DDX3 (Figure 6G). IKKɛ binding could provide a molecular explanation for the requirement of this DDX3 region for the effect on the Ifnb promoter.
To confirm the physiological relevance of the interaction between DDX3 and IKKɛ for PRR signalling, we infected HEK293T cells with SeV for different periods of time, and demonstrated a virus-induced transient association between endogenous DDX3 and endogenous IKKɛ (Figure 6H). This suggests that the detection of virus by a PRR induces the formation of a signalling complex containing IKKɛ and DDX3, thus providing further evidence implicating DDX3 in the pathway from PRRs to IRF activation and promoter induction.
Discussion
In this study, we demonstrate a novel function for DDX3 in innate immunity, namely as a positive regulator of TBK1/IKKɛ-mediated IRF activation. This novel function of DDX3 was revealed through the characterisation of the effect of the VACV protein K7 on host signalling pathways. Our interest in K7 stemmed from the fact that it shares significant sequence similarity with A52, a well-characterised inhibitor of the TLR–NF-κB signalling axis, which interacts with IRAK2 and TRAF6. As might be expected, K7 also targeted IRAK2 and TRAF6 and inhibited TLR-induced NF-κB activation. However, given the lack of effect of A52 on PRR pathways to IRF3 and 7 activation, it was surprising that K7 inhibited PRR signalling pathways to IRF3 and 7 activation, ISRE stimulation and Ifnb promoter induction.
We localised the inhibitory effect of K7 to TBK1/IKKɛ-mediated IRF activation, a point of convergence for many different PRR pathways that activate IRF3 and 7. Inhibiting TLR-independent pathways to Ifnb is likely to be relevant for VACV, a dsDNA virus that replicates in the cytoplasm and generates significant amounts of dsRNA (Weber et al, 2006). In fact, there is evidence that VACV engages TLR-independent pathways leading to IFN induction (Waibler et al, 2007; Zhu et al, 2007). Hence, it is also likely that VACV has developed counteracting strategies to evade these pathways and K7 appears to represent one such strategy. Given that TRAF6 and IRAK2 are not involved in TLR-independent IRF activation, we suspected that K7 targets an additional protein required for signalling to IRF activation, and subsequently identified DDX3 as such.
We provide several lines of evidence for a role for DDX3 in IRF activation: (1) DDX3 expression enhanced SeV-stimulated Ifnb promoter induction; (2) the N-terminal region of DDX3 was required for this effect and when expressed in isolation acted as a dominant negative to inhibit ligand-induced IRF3 and IRF3-dependent promoters; (3) knockdown of endogenous DDX3 expression by RNAi inhibited Ifnb promoter induction by TBK1 or IKKɛ, and SeV- or poly(I:C)-stimulated IRF3 activation; (4) IKKɛ interacted with the functionally important N-terminus of DDX3 and (5) The association of DDX3 with IKKɛ was inducible by SeV stimulation, linking DDX3 into the signalling pathway leading from RIG-I engagement to IRF activation.
The role of DDX3 in IRF activation in vivo remains to be confirmed by use of a DDX3 knockout mouse, as siRNA and classical transfection studies may not fully reflect the true in vivo role. Nonetheless, the virus-induced association between endogenous DDX3 and IKKɛ in human cells shown here strongly supports a signalling role for DDX3.
We also provide evidence that K7 targets this function of DDX3 to interfere with IRF activation, as DDX3 exerted its stimulatory effect at the level of TBK1/IKKɛ, which correlated with the point of inhibition by K7. Further, K7 bound to the N-terminal region of DDX3 that is required for Ifnb promoter induction. This is the first time a DEAD box helicase has been implicated in the regulation of an innate immune signalling pathway at the level of transcription factor activation by upstream kinases, although the distantly related DExD/H helicases RIG-I and mda-5 exert an effect further upstream as PRRs. Interestingly, DDX3 expression was shown to be induced in an IRF7-dependent manner (Barnes et al, 2004), an observation that further links DDX3 and the IRF pathway.
Prior to this study, an array of divergent cellular functions for human DDX3 and/or its orthologues have been reported, such as regulation of splicing and nuclear export of RNA (Rosner and Rinkevich, 2007). Recently, a positive role for DDX3 in cancer biogenesis has been suggested (Botlagunta et al, 2008). However, DDX3 had previously been proposed as a tumour suppressor due to its inhibitory effect on the translation initiation factor eIF4e (Shih et al, 2008). Still, DDX3 has been shown to upregulate p21 promoter activity by interacting with the Sp1 transcription factor and to enhance p21/waf1 protein expression (Chao et al, 2006). This latter finding is more in line with our discovery that DDX3 can upregulate Ifnb promoter induction through IRFs by interacting with IKKɛ. Interestingly, an emerging theme is that many DEAD box proteins are multifunctional and have roles in transcriptional regulation that are often independent of their helicase activity (Fuller-Pace, 2006). Consistent with this, we found that the positive effect of DDX3 on the Ifnb promoter is independent of its ATPase and unwinding activities. It is of note that the N-terminus of DDX3 is required for its role in IRF activation, as this region is very divergent in DEAD box helicases and is thought to confer functional specificity. It remains unclear how exactly the association of DDX3 and IKKɛ translates into enhanced IRF and promoter activation. The DDX3 N-terminus contains large numbers of serine clusters, and we observed a higher molecular weight form of DDX3 when co-expressed with IKKɛ. This might suggest that DDX3 is phosphorylated by IKKɛ. Other interesting features contained within the DDX3 N-terminal region are its nuclear export sequence (aa 11–21) and the recently described property to bind eIF4e (aa 38–44) (Shih et al, 2008). It is hence becoming clear that DDX3 has multiple different and distinct cellular functions involving different domains of DDX3, and our understanding of these functions is still fragmentary. Further research should investigate the interplay between the different functions and try to reconcile some of its seemingly contradictory effects.
Strikingly, other viral proteins also target DDX3: HIV Rev protein exploits the nuclear–cytoplasmic shuttling function of DDX3 to export virally encoded mRNAs out of the nucleus (Yedavalli et al, 2004). In contrast, VACV replicates in the cytoplasm and has therefore no requirement to subvert nuclear RNA export. It is unclear what the functional consequences of the HCV Core protein–DDX3 interaction are; however, it has recently been shown that DDX3 is required for HCV replication (Ariumi et al, 2007). K7 seems to target DDX3 in a different way compared to these two other viral proteins, namely by disrupting the ability of DDX3 to enhance IRF activation by binding the functionally important N terminus. Neither Rev nor the Core protein binds to DDX3 within the N-terminal region. Also, HIV and HCV seem to subvert rather than inhibit DDX3, considering that DDX3 is required for HIV and HCV replication. It is possible that by recruiting DDX3 for the purpose of nuclear export of viral mRNAs and/or viral replication, these viruses simultaneously sequester DDX3 away from its positive effector role in the IRF pathway and are hence benefiting in two different ways from targeting DDX3.
Because of its requirement for HIV and HCV replication, DDX3 has been suggested as an antiviral drug target, as inhibition of DDX3 might block replication of these viruses (Kwong et al, 2005). However, in the light of our findings this strategy should be carefully assessed, as disruption of DDX3 would also impair the induction of antiviral mediators that depend on IRF activation. In conclusion, we have shown a novel positive role for a DEAD box protein in IRF activation by TBK1/IKKɛ. This study should further the interest in DDX3 by drawing attention to its novel role in innate immunity. More in-depth studies of how DDX3 regulates IRF activation and of the mechanism whereby K7 inhibits DDX3 function are underway and should provide us with valuable information for manipulating DDX3 function in the future.
Materials and methods
Plasmids
Sources of expression plasmids were Flag–TRAF6 and Flag–TRAF2 (Tularik Inc., San Francisco, CA), CD4-TLR4 (R Medzhitov, Yale University, CT), Flag–TRIF (S Akira, Osaka University, Osaka, Japan), Myc–MyD88 (L O'Neill, Trinity College Dublin, Dublin, Ireland), TBK1–Flag, IKKɛ–Flag (K Fitzgerald, University of Massachusetts Medical School, Worcester, MA) and MAVS (Z Chen, University of Texas, TX). K7R was amplified from genomic DNA of the VACV strain WR and the PCR product was cloned into pCMV-HA to make pCMV-HA-K7R. HA–K7R was also subcloned into pRK5 mammalian expression vector. For bacterial expression of His-tagged K7, K7R was subcloned into pHisParallel2. DDX3 was amplified from human PBMC cDNA and the PCR product was cloned into the pCMV-Myc and pCMV-HA mammalian expression vectors (BD Biosciences) and into pHisParallel2 for bacterial expression of His-tagged DDX3. For generating K7–EYFP fusion protein, K7R was cloned into pCDNA (Invitrogen) containing the EYFP ORF (provided by K Kroeger, WAIMR, Perth, Australia).
Antibodies and reagents
Anti-K7 polyclonal Ab was raised against purified full-length recombinant K7 expressed from pHisparallel2-K7R in Escherichia coli (Inbiolabs). Other antibodies used were anti-Flag M2mAb, anti-Flag M2-conjugated agarose, anti-Myc mAb clone 9E10 (all from Sigma), anti-HA mAb (Covance), anti-DDX3 clone BL1649 (Bethyl Laboratories, Montgomery, TX, USA), anti-IKKɛ (Abcam, Cambridge, UK), anti-IRF3 (IBL) anti-phospho-Ser396-IRF3 (Cell Signaling Technologies) and anti-A52 (Harte et al, 2003). Human rIL-1β was a gift from the National Cancer Institute (Frederick, WA). Poly(I:C) and poly(dA:dT) were from Amersham. SeV was kindly provided by Ilkka Julkunen (National Public Health Institute, Finland).
Reporter gene assays
Promoter induction and transcription factor activation were measured using HEK293 cells (2 × 104 cells per well) seeded onto 96-well plates and transfected 24 h later with expression vectors and luciferase reporter genes (Maloney et al, 2005; Stack et al, 2005). Data are expressed as the mean fold induction±s.d. relative to control levels, for an individual experiment performed in triplicate. A representative of at least three independent experiments is shown.
Immunoprecipitation and immunoblotting
Co-immunoprecipitations were performed in HEK293T cells (1.5 × 106 cells) seeded in 10-cm dishes (Stack et al, 2005). The immune complexes were analysed by SDS–PAGE and immunoblotting. For analysis of the kinetics of K7 expression, confluent monolayers of HEK293 cells were infected at 10 p.f.u. VACV per cell for 60 min at 37°C.
His pull-down assays
Bacterial expression constructs were transformed into E. coli BL21 (DE3). Protein expression was induced with IPTG, and proteins were purified using Ni-Agarose (Sigma). For pull-down experiments, equal amounts of the different His fusion proteins were used as determined by SDS–PAGE and Coomassie staining. Cell lysate from transfected HEK293 cells was incubated with the purified His fusion proteins coupled to Ni-Agarose. The complexes were precipitated and subjected to SDS–PAGE. For the identification of K7-interacting proteins, specific bands were cut out of the Coomassie-stained gel and prepared for MALDI-TOF analysis. For all other pull-down experiments, gels were transferred to PVDF membranes and subjected to immunoblotting.
MALDI-TOF
Bands were cut from Coomassie-stained SDS–PAGE gels and digested with 10 μg/ml modified trypsin (Promega) overnight at 37°C. Peptides were extracted and samples were analysed by MALDI-TOF. The resulting peak list was searched against the NCBI non-redundant protein database using the Mascot search engine. DDX3 was identified with a probability-based Mowse score of 120.
Determination of cytokine concentrations
LPS-induced IL-8 and poly(I:C)-induced RANTES production in HEK293 clonal cell lines expressing either TLR4 and MD-2 (HEK-TLR4) or TLR3 (HEK-TLR3) was measured by ELISA.
Confocal imaging
HEK293 cells were grown on 22-mm coverslips in six-well plates and transfected with 2.3 μg of total DNA. At 48 h after transfection, cells were fixed with 4% paraformaldeyde, permeabilised with 0.5% Triton-X-100 and stained with anti-HA-AlexaFluor594 and DAPI nuclear stain. Slides were examined by phase-contrast and confocal microscopy with an Olympus Fluoview FV1000 Imaging system using a × 60 objective and 2- to 3-fold digital zoom. Images were collected with Kalman integration and processed with the Olympus Fluoview software (Version 1.3c).
siRNA gene silencing
For silencing of endogenous human DDX3, two different Stealth™ RNAi oligonucleotides were employed: DDX3-1 5′-GGGAGAAAUUAUCAUGGGAAACAUU-3′ (HSS102712) and DDX3-2 5′-UUCAACAAGAAGAUCCAACAAAUCC-3′ (HSS102713) along with a control oligonucleotide of a matched GC content (36% GC) (Invitrogen). HEK293 cells were transfected with the RNAi duplexes using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. This transfection was repeated after 24 h and cells were harvested 24 h after the second transfection. The cell lysates were prepared and analysed by SDS–PAGE and immunoblotting. For reporter gene assays, HEK293 cells in 96-well plates (2 × 104 cells per well) were transfected with siRNA duplexes and then co-transfected with reporter and expression constructs (or stimulated with SeV or poly(I:C)) and a second dose of siRNA 24 h later. Reporter gene induction was analysed 24 h after the second transfection.
Acknowledgments
We thank Caroline Jefferies for help with MALDI-TOF and peptide mass fingerprinting, and Orla Hanrahan for technical support. This study was supported by Science Foundation Ireland and the Health Research Board (Career Development Fellowship to MS).
References
- Ariumi Y, Kuroki M, Abe K, Dansako H, Ikeda M, Wakita T, Kato N (2007) DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication. J Virol 81: 13922–13926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes BJ, Richards J, Mancl M, Hanash S, Beretta L, Pitha PM (2004) Global and distinct targets of IRF-5 and IRF-7 during innate response to viral infection. J Biol Chem 279: 45194–45207 [DOI] [PubMed] [Google Scholar]
- Botlagunta M, Vesuna F, Mironchik Y, Raman A, Lisok A, Winnard P Jr, Mukadam S, Van Diest P, Chen JH, Farabaugh P, Patel AH, Raman V (2008) Oncogenic role of DDX3 in breast cancer biogenesis. Oncogene 27: 3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie A, Kiss-Toth E, Symons JA, Smith GL, Dower SK, O'Neill LA (2000) A46R and A52R from vaccinia virus are antagonists of host IL-1 and Toll-like receptor signaling. Proc Natl Acad Sci USA 97: 10162–10167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie AG (2007) Translational mini-review series on Toll-like receptors: recent advances in understanding the role of Toll-like receptors in anti-viral immunity. Clin Exp Immunol 147: 217–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao CH, Chen CM, Cheng PL, Shih JW, Tsou AP, Lee YH (2006) DDX3, a DEAD box RNA helicase with tumor growth-suppressive property and transcriptional regulation activity of the p21waf1/cip1 promoter, is a candidate tumor suppressor. Cancer Res 66: 6579–6588 [DOI] [PubMed] [Google Scholar]
- Fuller-Pace FV (2006) DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res 34: 4206–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harte MT, Haga IR, Maloney G, Gray P, Reading PC, Bartlett NW, Smith GL, Bowie A, O'Neill LA (2003) The poxvirus protein A52R targets Toll-like receptor signaling complexes to suppress host defense. J Exp Med 197: 343–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogbom M, Collins R, van den Berg S, Jenvert RM, Karlberg T, Kotenyova T, Flores A, Karlsson Hedestam GB, Schiavone LH (2007) Crystal structure of conserved domains 1 and 2 of the human DEAD-box helicase DDX3X in complex with the mononucleotide AMP. J Mol Biol 372: 150–159 [DOI] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G (2006) 5′-Triphosphate RNA is the ligand for RIG-I. Science 314: 994–997 [DOI] [PubMed] [Google Scholar]
- Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, Uematsu S, Takeuchi O, Takeshita F, Coban C, Akira S (2008) TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature 451: 725–729 [DOI] [PubMed] [Google Scholar]
- Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, Akira S (2005) Cell type-specific involvement of RIG-I in antiviral response. Immunity 23: 19–28 [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S (2006) Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441: 101–105 [DOI] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S (2005) IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol 6: 981–988 [DOI] [PubMed] [Google Scholar]
- Keating SE, Maloney GM, Moran EM, Bowie AG (2007) IRAK-2 participates in multiple Toll-like receptor signaling pathways to NF{kappa}B via activation of TRAF6 ubiquitination. J Biol Chem 282: 33435–33443 [DOI] [PubMed] [Google Scholar]
- Kwong AD, Rao BG, Jeang KT (2005) Viral and cellular RNA helicases as antiviral targets. Nat Rev Drug Discov 4: 845–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney G, Schroder M, Bowie AG (2005) Vaccinia virus protein A52R activates p38 mitogen-activated protein kinase and potentiates lipopolysaccharide-induced interleukin-10. J Biol Chem 280: 30838–30844 [DOI] [PubMed] [Google Scholar]
- Mamiya N, Worman HJ (1999) Hepatitis C virus core protein binds to a DEAD box RNA helicase. J Biol Chem 274: 15751–15756 [DOI] [PubMed] [Google Scholar]
- McWhirter SM, Fitzgerald KA, Rosains J, Rowe DC, Golenbock DT, Maniatis T (2004) IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc Natl Acad Sci USA 101: 233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J (2005) Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437: 1167–1172 [DOI] [PubMed] [Google Scholar]
- Moynagh PN (2005) TLR signalling and activation of IRFs: revisiting old friends from the NF-kappaB pathway. Trends Immunol 26: 469–476 [DOI] [PubMed] [Google Scholar]
- O'Neill LA (2006) How Toll-like receptors signal: what we know and what we don't know. Curr Opin Immunol 18: 3–9 [DOI] [PubMed] [Google Scholar]
- Owsianka AM, Patel AH (1999) Hepatitis C virus core protein interacts with a human DEAD box protein DDX3. Virology 257: 330–340 [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C (2006) RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314: 997–1001 [DOI] [PubMed] [Google Scholar]
- Rosner A, Rinkevich B (2007) The DDX3 subfamily of the DEAD box helicases: divergent roles as unveiled by studying different organisms and in vitro assays. Curr Med Chem 14: 2517–2525 [DOI] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ (2005) Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122: 669–682 [DOI] [PubMed] [Google Scholar]
- Shih JW, Tsai TY, Chao CH, Wu Lee YH (2008) Candidate tumor suppressor DDX3 RNA helicase specifically represses cap-dependent translation by acting as an eIF4E inhibitory protein. Oncogene 27: 700–714 [DOI] [PubMed] [Google Scholar]
- Stack J, Haga IR, Schroder M, Bartlett NW, Maloney G, Reading PC, Fitzgerald KA, Smith GL, Bowie AG (2005) Vaccinia virus protein A46R targets multiple Toll-like-interleukin-1 receptor adaptors and contributes to virulence. J Exp Med 201: 1007–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka A, Taniguchi T (2007) Cytosolic DNA recognition for triggering innate immune responses. Adv Drug Deliv Rev 31: 31. [DOI] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, Taniguchi T (2007) DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448: 501–505 [DOI] [PubMed] [Google Scholar]
- Waibler Z, Anzaghe M, Ludwig H, Akira S, Weiss S, Sutter G, Kalinke U (2007) Modified vaccinia virus Ankara induces Toll-like receptor-independent type I interferon responses. J Virol 81: 12102–12110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR (2006) Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol 80: 5059–5064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB (2005) VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell 19: 727–740 [DOI] [PubMed] [Google Scholar]
- Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT (2004) Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 119: 381–392 [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T (2004) The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5: 730–737 [DOI] [PubMed] [Google Scholar]
- You LR, Chen CM, Yeh TS, Tsai TY, Mai RT, Lin CH, Lee YH (1999) Hepatitis C virus core protein interacts with cellular putative RNA helicase. J Virol 73: 2841–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Martinez J, Huang X, Yang Y (2007) Innate immunity against vaccinia virus is mediated by TLR2 and requires TLR-independent production of IFN-beta. Blood 109: 619–625 [DOI] [PMC free article] [PubMed] [Google Scholar]