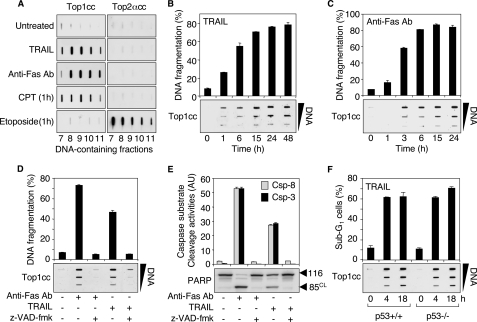
FIGURE 1.
TRAIL and Fas induce apoptotic Top1cc. A, TRAIL (0.1 μg/ml, 48 h) and anti-Fas Ab (0.1 μg/ml, 48 h) induce Top1cc but fail to form Top2cc in Jurkat cells. CPT (1 μm, 1 h) and etoposide (100 μm, 1 h) were used as positive controls for Top1cc and Top2cc, respectively. The DNA-containing fractions were probed by immunoblotting with antibodies against Top1 and Top2α as indicated. B and C, the kinetics of formation of Top1cc parallel TRAIL- and Fas-induced DNA fragmentation. Jurkat cells were treated with 0.1 μg/ml TRAIL (B) or 0.1 μg/ml anti-Fas Ab (C) for the indicated times. Top panels, apoptotic DNA fragmentation (means ± S.D. of triplicate samples). Bottom panels, detection of Top1cc. The DNA-containing fractions were pooled and probed at three concentrations (10, 3, and 1 μg) with an antibody against Top1. D and E, the pan-caspase inhibitor Z-VAD-fmk inhibits TRAIL- and Fas-induced apoptotic Top1cc. Jurkat cells were treated with Z-VAD-fmk (100 μm, 30 min) before the addition of anti-Fas Ab (0.1 μg/ml) or TRAIL (0.1 μg/ml) for 6 h. D, top panel, apoptotic DNA fragmentation (means ± S.D. of triplicate samples). Bottom panel, detection of Top1cc as for B and C. E, top panel, IETD-AFC, (caspase-8 (Csp-8), gray bars) and DEVD-AFC (caspase-3 (Csp-3), black bars) peptide cleavage activity (means ± S.D. of triplicate samples). Bottom panel, whole cell extracts were examined for PARP cleavage by Western blotting. Molecular mass of PARP polypeptide and its cleaved product (CL) is indicated at right. F, TRAIL induces apoptotic Top1cc independently of p53. HCT116 cells of each genotype (p53+/+ and p53-/-) were treated with 0.1 μg/ml TRAIL for the indicated times. Top panel, percentages of cells with apoptotic sub-G1 DNA (means ± S.D. of triplicate samples). Bottom panel, detection of Top1cc as for B and C.