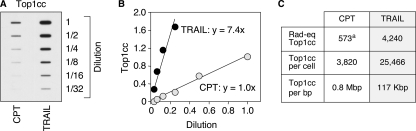
FIGURE 5.
Quantification of the apoptotic Top1cc induced by TRAIL. A, HCT116 cells were treated with 0.1 μg/ml TRAIL for 4 h. Top1cc were quantitated by pooling the DNA-containing fractions and probing with an antibody against Top1. The pools containing 10 μg of DNA were assayed by sequential 2-fold dilutions. HCT116 cells treated by CPT (0.5 μm, 1 h), for which the number of Top1cc/cell has been determined (39), were used to determine the genomic frequency of apoptotic Top1cc. B, quantification of the data shown in A. The curves displaying the levels of Top1cc as a function of the dilution of the samples show a 7.4-fold increased of Top1cc induced by TRAIL as compared with CPT (0.5 μm). C, table expressing the amount of rad equivalent (Rad-eq) Top1cc, and the number of Top1cc per cell and per bp in response to TRAIL. a, CPT (0.5 μm, 1 h) induces 573 rad equivalent Top1cc in HCT116 cells (39), and 1 rad equivalent Top1cc corresponds to ∼1 Top1cc/0.9 × 109 nucleotides (64). The values for the TRAIL-treated cells were obtained by multiplying by 7.4 (see B) those of the standard CPT.