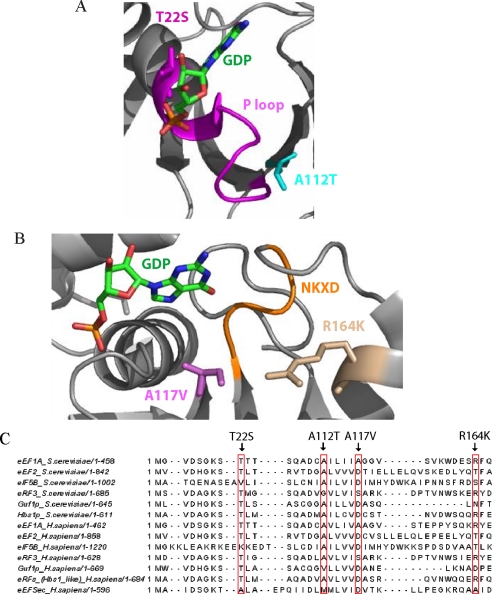
FIGURE 7.
GEF-independent mutants of eEF1A either affect the P-loop or the NKXD element. A, Thr-22 (magenta) and Ala-112 (cyan) residues and their positions respective to the nucleotide (green) and the P-loop (magenta) are shown. B, Arg-164 (wheat) and Ala-117 (purple) residues and their positions respective to the nucleotide (green) and the NKXD motif (yellow) are shown. The structure was produced with the PyMOL program (32), using the co-crystal structure of the eEF1A·eEF1Bα complex with GDP (Protein Data Bank code 1IJE) (39). C, sequence alignment of yeast eEF1A with the G-proteins independent of GEF in yeast (eEF2, eIF5B, eRF3, Guf1p, and Hbs1p) and human (eEF2, eIF5B, eRF3, Guf1p, Hbs1-like eRFs, and eEFSec). The alignment was generated by Jalview (41).