Figure 1. Sequence and structure of CaM-binding peptides.
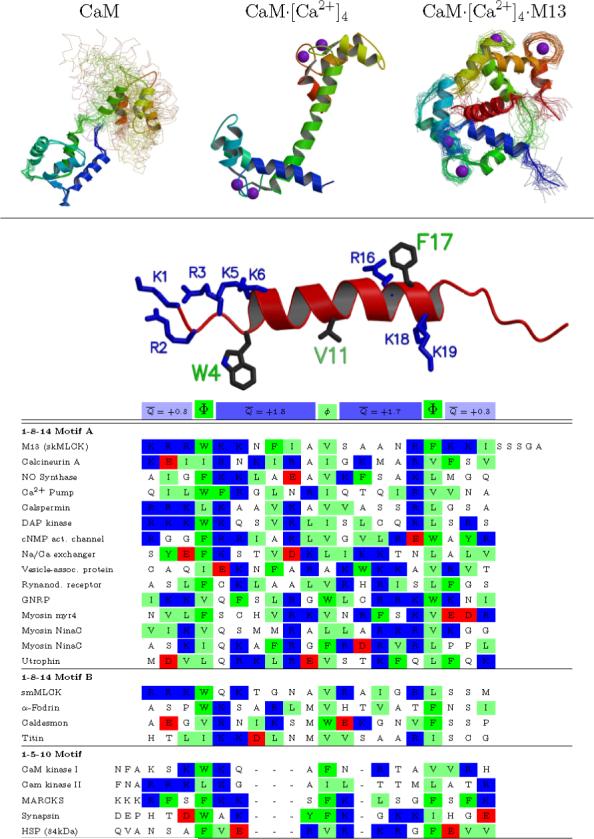
(A) CaM: The solution structure of CaM in the absence of Ca2+ shows relatively well structured N- and C-terminal domains but free motion between the domains. CaM·[Ca2+]4: The crystal structure of CaM in the presence of calcium shows a different structure for each domain, and a single well-defined helical connection between them (although this region is flexible in solution). CaM·[Ca2+]4·M13: The solution structure of peptide-bound CaM shows a domain structure similar to CaM·[Ca2+]4, but a very different relation between them —the linking helix bends to enclose the helical peptide between the two domains. (B) An alignment of a representative set of mammalian CaM-binding peptides shows a general motif, but no clear consensus sequence. Two larger hydrophobic residues are spaced 2.5 or 3.5 helical turns apart, and anchor the peptide into each domain of CaM. An additional smaller hydrophobic group located one or two turns from the first is also conserved. There is no further conservation at any given position, but all the peptides are basic (+1 to +8 net charge). This net charge is fairly evenly distributed, with an average of roughly +1.5e located in each of the regions between the hydrophobic anchors, and an average of roughly +0.5e on the three residues on either side of the of the core helical region. Sequences taken from Rhoads and Friedberg. (4) Structural figure generated with molscript (60) and raster3d. (61)
