Figure 4. Charge reversal at M13 site 19.
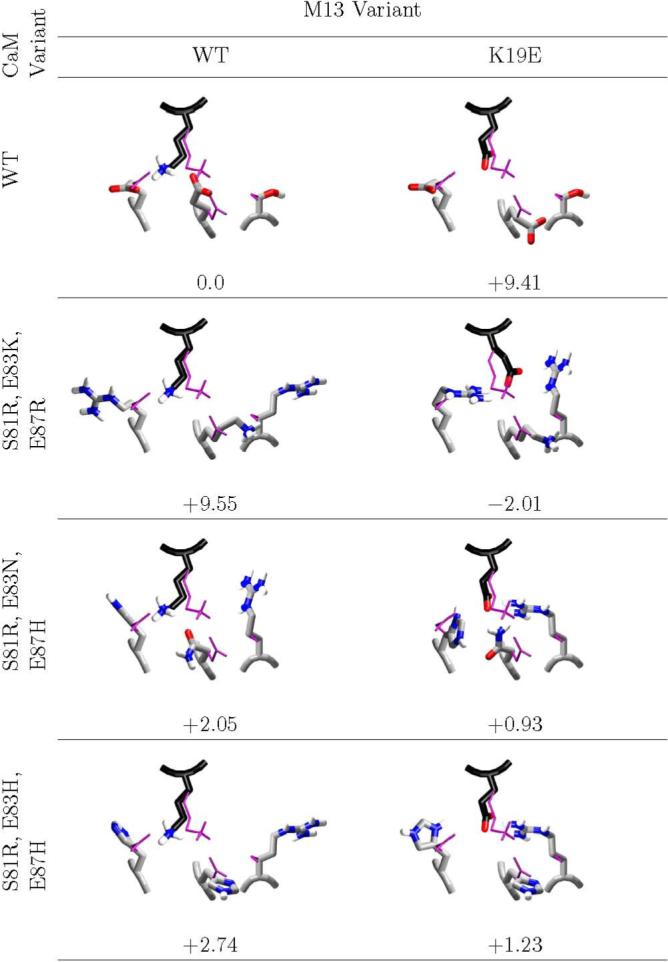
The computed minimum energy complex structures of design candidates and decoys at M13 position 19 are shown, along with the computed binding affinity, relative to wild-type. The desired complexes have computed affinities ranging from 2.0 kcal/mol more stable than wild-type to 1.2 kcal/mol destablized; all decoys are computed to be destabilized by greater than 2.0 kcal/mol. In all cases, the wild-type NMR average structure is shown in thin purple. Carbons of M13 are colored black, and carbons of CaM are colored light grey. Structural figure generated with vmd (62) and raster3d. (61)
