Figure 6.
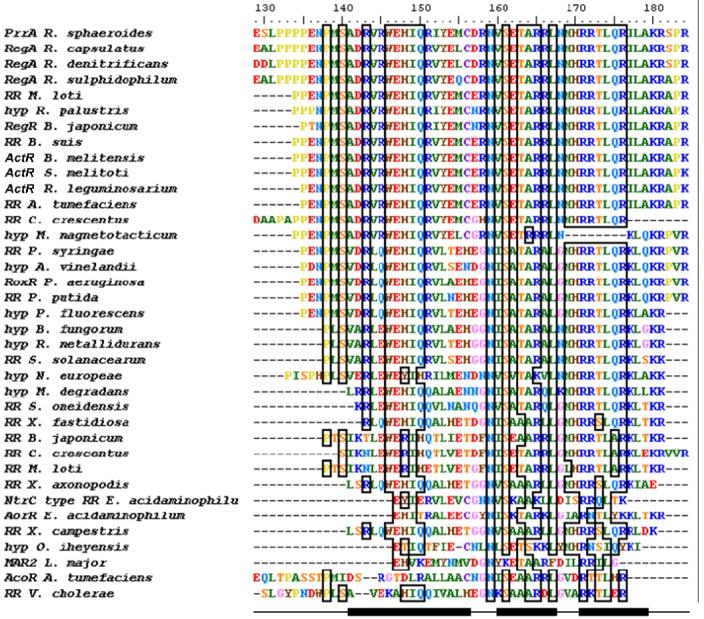
Alignment of PrrAC with homologous RR effector domains and hypothetical proteins. Conservation >70% is outlined by boxes. PrrAC secondary structure (helices α6-8) is indicated at the bottom of the figure. The conservation occurs mainly in the first loop and the helix-turn-helix motif. Residues shown to be involved in DNA binding in the first loop (binding to phosphate backbone) and at the beginning of α8 (specific binding to bases) are highly conserved. Hyp: hypothetical protein. Species not otherwise given in the text are: Mesorhizobium loti, Rhodopseudomonas loti, Brucella suis, Brucella melitensis, Rhizobium leguminosarium, Agrobacterium tumefaciens, Caulobacter crescentus, Magnetospirillum magnetotacticum, Azotobacter vinelandii, Ralstonia metallidurans, Ralstonia solanacearum, Microbulbifer degradans, Shewanella oneidensis, Xylella fastidiosa, Eubacterium acidaminophilum, Xanthomonas campestris, Oceanobacillus iheyensis, Leishmania major.
