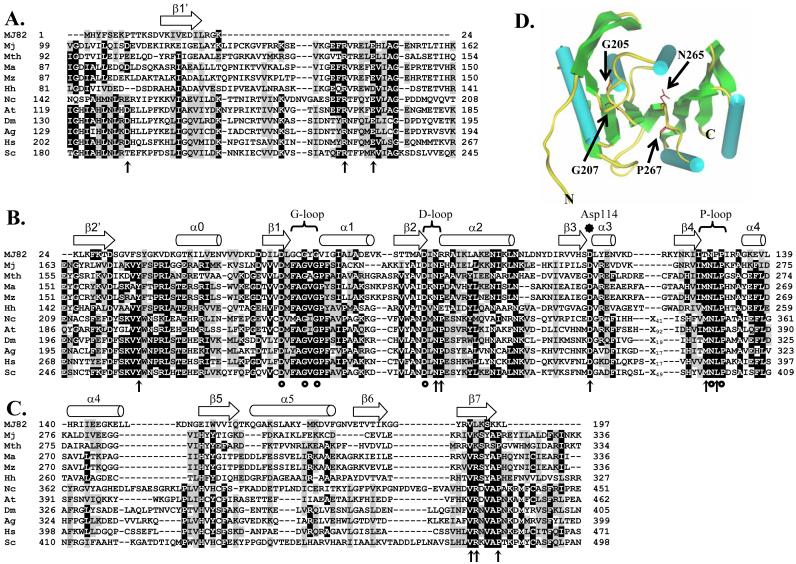
Fig.1.
Sequence alignment of archaea and eukaryotic Trm5 proteins by ClustalW, together with modeling of the alignment based on the crystal structure of MJ0882 (25). Significance scores are not calculated for this alignment. Sequences used in the alignment include Trm 5 of M. jannaschii (Mj), Methanobacterium thermoautotrophicum (Mth), M. acetivorans (Ma), M. mazei (Mz), Halobacterium halobium (Hh), Neurospora crassa (Nc), Arabidopsis thalania (At), Drosophila melanogaster (Dm), Anopheles gambiae (Ag), Homo sapiens (Hs), and Saccharomyces cerevisiae (Sc). Characteristic secondary structural elements α-helices and β-strands are indicated by arrows and cylinders, respectively, across the top of the alignment. Mutations that had been previously published are indicated by circles, while those that were created in this study are indicated by vertical arrows. Trm 5 is divided into three domains: (A) the N-terminal domain, (B) the AdoMet-binding domain, and (C) the C-terminal domain. In (D), crystal structure of MJ0882 is shown, where cylinders and arrows represent α-helices and β-strands, respectively, according to the published assignments in the crystal structure (25). No additional modeling is performed on the structure of MJ0882, thus no ProsaII or Verify3D is employed. The positions of G205, G207, N265, and P267 residues of M. jannaschii Trm5 are deduced from the sequence alignment with MJ0882, and are shown in red, indicating their positions in the AdoMet-binding site.