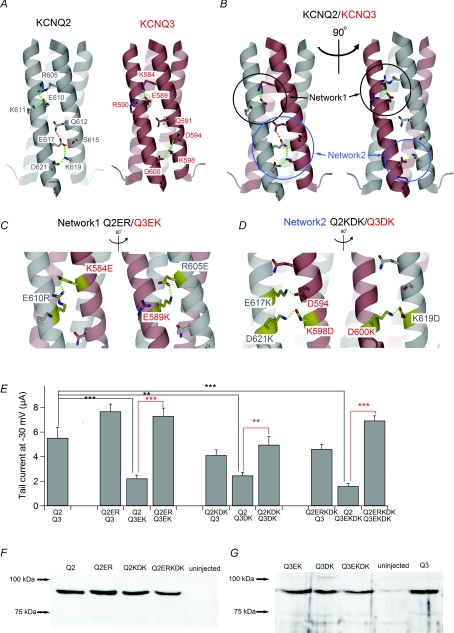
Figure 5. Salt bridge networks in TCC2 are important for subunit recognition and efficient current expression.
A, speculated structures of TCC2 in homomeric KCNQ2 and KCNQ3 channels based on the crystallographic structure of KCNQ4 TCC2 (see Methods). Green and orange dotted lines represent deduced salt bridges and hydrogen bonds, respectively. Amino acid residues involved in the salt bridge/hydrogen bond network are labelled. B, speculated TCC2 structure in the heteromeric KCNQ2/3 channel. KCNQ2/3 can have two different sides having different sets of electrostatic contacts, and both are shown. Green and orange dotted lines represent putative salt bridges and hydrogen bonds, respectively. Networks 1 and 2 are circled in black and blue, respectively. C and D, speculated structures for Networks 1 (C) and 2 (D) within different combinations of charge-swapped mutants. Two possible sides are shown for each. Green dotted lines represent putative salt bridges. Mutated amino acid residues are coloured yellow and labelled. Endogenous D594 in KCNQ3 is also labelled because it can make a salt bridge with E617K in the Q2KDK mutant, as depicted in D. All other salt bridges are formed between mutated residues. E, multiple comparisons of maximum tail current amplitudes among charge-swapped mutants. **P < 0.01, ***P < 0.001 (Tukey's test). F, expression levels for the charge-swapped mutants of KCNQ2 analysed by immunoblotting with anti-KCNQ2 antibody. KCNQ3 was not coexpressed. G, expression levels for the charge-swapped mutants of KCNQ3 analysed by immunoblotting with anti-KCNQ3 antibody. KCNQ2 was not coexpressed.