Fig. 5.
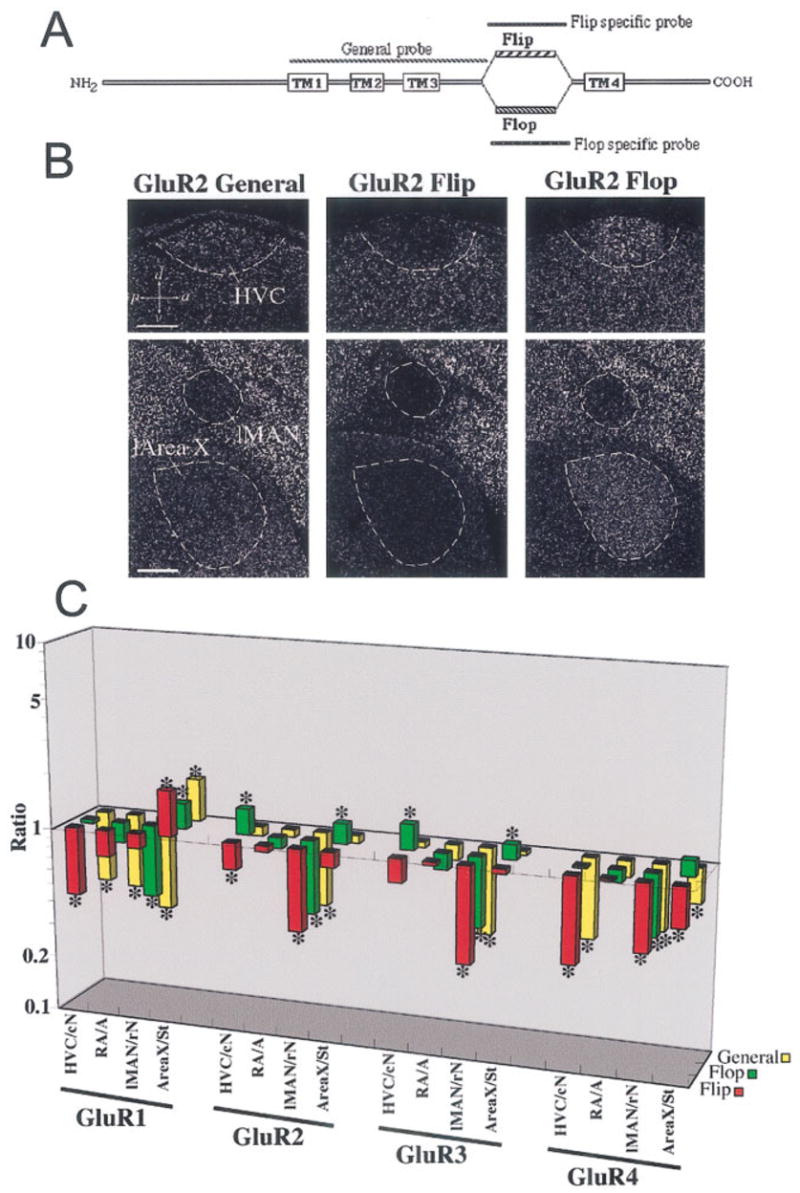
Differential regulation of mRNA splicing of AMPA subunits in vocal nuclei. A: General schematic of mammal AMPA receptor proteins with their four transmembrane domains (TM1–TM4). Between TM3 and TM4 are the alternatively spliced extracellular domains Flip or Flop. The general probe hybridizes to the TM1–TM3 region of both splice variant mRNAs. Each specific probe hybridizes to the Flip or Flop spliced form only. B: Example of differences in expression of AMPA receptor splice variants revealed by comparing hybridizations with the GluR2 general, Flip, and Flop probes in zebra finch vocal nuclei. There is lower expression of GluR2 Flip and higher expression of GluR2 Flop mRNA in HVC and l-Area X compared with the surrounding brain subdivision. C: Quantification of differential expression of splice variants of all four AMPA subunits (GluR1–4; gene expression level on the z-axis, brain region on the x-axis, and gene on the y-axis). The z-axis is log scaled to view comparable ratios above and below 1 graphically. Further explanation of ratio interpretations is given in the legend to Figure 4. *P < 0.01 (Fisher’s PLSD test; n = 3 male zebra finches). Scale bars = 300 μm.
