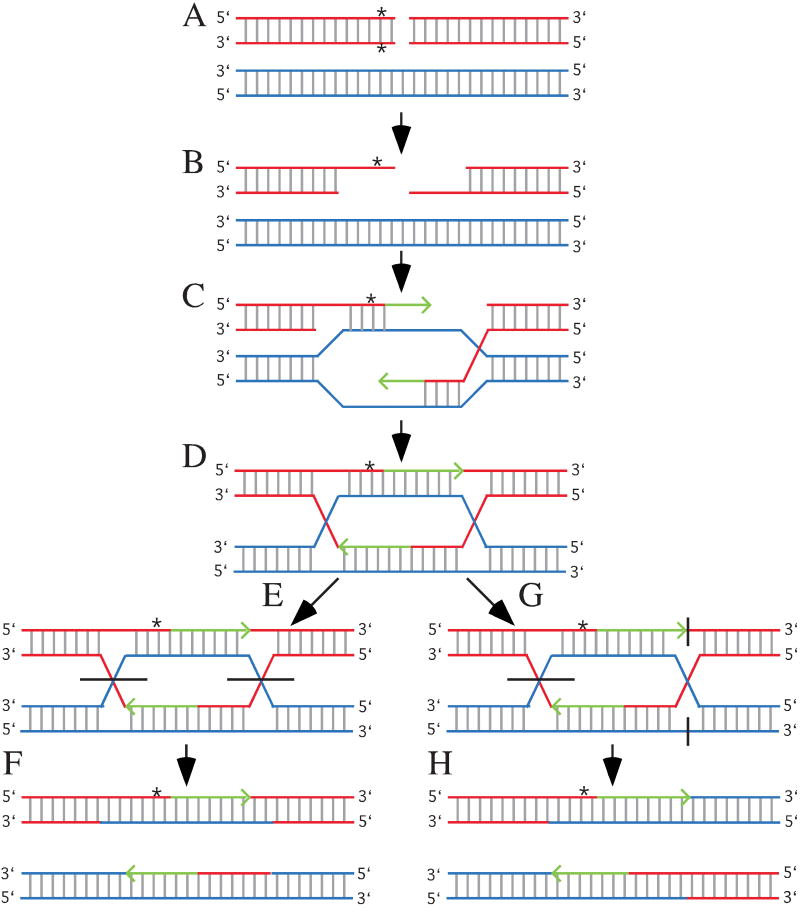
Fig. 1.
Homologous recombination and gene conversion. (a) Homologous templates are aligned. This usually occurs between sister chromatids during DNA replication, but may also occur between chromosomes as shown here. The red chromosome has a DSB and a sequence difference (*) located close to the break. (b) The ends are resected to form single-strands. Note the sequence difference from the bottom red strand is removed. (c) The red single-strands invade the blue homologous template to form a joint molecule and primers for DNA replication (green arrows). Note the blue strands are used as templates for DNA replication; therefore, the green strands contain the same information as the blue strands. This means sequence differences on the resected red strand are lost (gene conversion). (d) The formation of Holliday junctions (a mobile attachment between four strands of DNA) and heteroduplexes (double-strand DNA composed of a strand from each homologue). Note the sequence difference is lost for one strand and a mismatch is formed for the other strand. (e) Noncrossover resolution of the Holliday junctions. Horizontal black lines represent locations where strands are cut. (f) The final products. For this diagram the top chromosome contains a mismatched that may be resolved by MMR. (g) Crossover resolution. The left Holliday junction is resolved as shown for the noncrossover event; however, the outer strands are cut to resolve the right Holliday junction. (g) The final products showing the chromosomes recombine. The heteroduplex and mismatch are the same as described for the noncrossover.