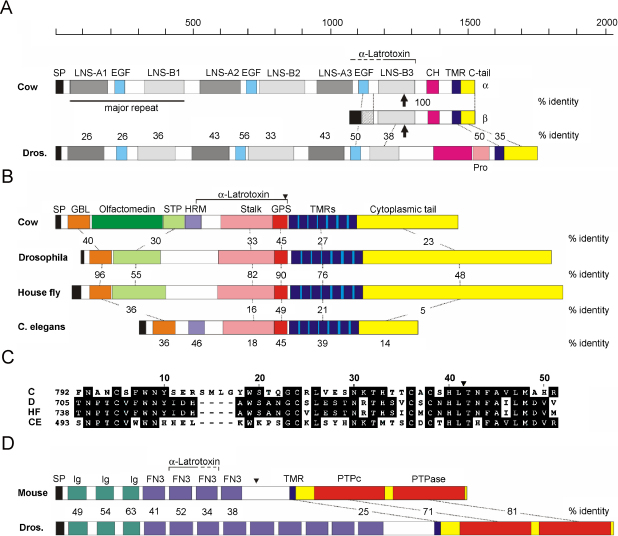
Fig. 4.
Evolutionary conservation of the three types of α-LTX receptors. (A) Neurexins in cow and Drosophila. Vertebrates possess three neurexin genes, each of which encodes a long (α) and a short (β) form of neurexin. (B) Three latrophilins are found in vertebrates (LPH1 from the cow, Bos taurus, is shown), whereas only one form is present in insects (Drosophila and house fly, M. domestica). The nematode (C. elegans) LPH is illustrated for comparison because it binds ε-LIT from Latrodectus venom (Mee et al., 2004). (C) Multiple sequence alignment of the GPS domains from cow, C; Drosophila, D; housefly, HF; and C. elegans, CE. Identical residues are highlighted. (D) Receptor-like protein tyrosine phosphatase σ (PTPσ) in mouse and Drosophila. (A–D) Percentage identities are indicated by numbers between the respective domains (values below the C. elegans structure correspond to the cow sequence). The minimal protein regions required for α-LTX binding are indicated by square brackets above the diagrams. Domain names are abbreviated as follows: CH, O-linked carbohydrate domain; EGF, epidermal growth factor-like domains; FN3, fibronectin III-like domain; GBL, galactose-binding lectin-like domain; GPS, G-protein-coupled receptor proteolysis site; HRM, hormone receptor motif; Ig, immunoglobulin-like domain; LNS, laminin G-domain/neurexin/sex hormone binding protein repeat; SP, signal peptide; STP, serine/threonine/proline-rich region; Pro, proline-rich region; PTPase, protein tyrosine phosphatase; TMR, transmembrane region. Black arrows in A denote the site of alternative splicing #4 within the last LNS repeat; small black arrowheads in B and D indicate the sites of constitutive proteolysis.