Abstract
Background
Bacteria use N-acyl homoserine lactone (AHL) molecules to regulate the expression of genes in a density-dependent manner. Several biosensors have been developed and engineered to detect the presence of all types of AHLs.
Results
In this study, we describe the usefulness of a traI-luxCDABE-based biosensor to quickly detect AHLs from previously characterized mutants of Burkholderia cenocepacia and Pseudomonas aeruginosa in both liquid and soft-agar co-culture assays in a high-throughput manner. The technique uses a co-culture system where the strain producing the AHLs is grown simultaneously with the reporter strain. Use of this assay in liquid co-culture allows the measurement of AHL activity in real time over growth. We tested this assay with Burkholderia cenocepacia and Pseudomonas aeruginosa but it should be applicable to a broad range of gram negative species that produce AHLs.
Conclusion
The co-culture assays described enable the detection of AHL production in both P. aeruginosa and B. cenocepacia and should be applicable to AHL analysis in other bacterial species. The high-throughput adaptation of the liquid co-culture assay could facilitate the screening of large libraries for the identification of mutants or compounds that block the synthesis or activity of AHLs.
Background
Bacteria possess various regulatory systems that enable them to quickly adapt to subtle changes in their environment. Variation in population density is a condition that bacteria are capable of perceiving and responding to in order to coordinate a myriad of behaviors, often referred as quorum sensing (QS) [1]. The molecular basis of most QS-dependent systems in Gram-negative bacteria is similar to the LuxI/R system of Vibrio fischeri, well known for the control of bioluminescence [2]. Briefly, a luxI homologue encodes for a N-acyl homoserine lactone (AHL) synthase that catalyzes the synthesis of AHL signal molecules, and a luxR homologue encodes a sensor/response regulator, which binds its cognate AHL in order to regulate the expression of genes usually in a density-dependent manner [3]. Recognition of cognate AHLs is dependent on the length of the acyl-chain and position C3 which can be either unmodified or carry an oxo- or hydroxyl group substitution.
Several biosensor systems have been developed that vary with respect to sensitivity and specificity for the detection of the AHL signals (reviewed in [4]). Recently, our laboratory developed a new bacterial biosensor using the TraI/R system of Agrobacterium tumefaciens to first detect the presence of AHLs in the lungs of rats and mice infected with Burkholderia cenocepacia [5] and subsequently from mucopurulent secretions of cystic fibrosis patients infected with Pseudomonas aeruginosa and/or B. cepacia complex organisms [6]. This biosensor system was engineered in the strain A. tumefaciens A136 cured of the Ti plasmid [7] to eliminate the TraI/R QS system. Our reporter strain carries two plasmids, pCF218, which constitutively produces the TraR response regulator [8] and pMV26 [5,6], which contains the traI promoter fused to the luxCDABE operon [9]. TraR binds AHLs present in the same environment as the reporter strain resulting in an AHL-TraR complex that subsequently binds to a specific sequence within the traI promoter on pMV26, triggering the transcription of the luxCDABE operon and the production of bioluminescence. This biosensor was shown to respond to AHLs with acyl side chains ranging from 4 to 12 carbons with greater sensitivity to AHLs with longer side chains and those with 3-oxo-substitutions [6].
In the present study, we describe the use of the traI-luxCDABE fusion [5,6] as a powerful bacterial biosensor for the in vitro detection of AHLs in co-culture assays in both liquid and semi-solid agar and its application as a high-throughput screening tool using B. cenocepacia and P. aeruginosa, two species with well characterized quorum sensing systems. B. cenocepacia has two QS systems, designated CepIR and CciIR that regulate genes involved in virulence, biofilm formation, swarming motility, and regulation [10]. B. cenocepacia strain K56-2 produces two AHLs with N-octanoyl-l-HSL (C8-HSL) synthesized primarily by the cepI gene product and N-hexanoyl-l-HSL (C6-HSL) synthesized mainly via the CciI synthase [11-13]. P. aeruginosa has two QS systems, designated LasIR and RhlIR, that influence the expression of approximately 5% of the genome including genes involved in virulence and biofilm formation [14]. P. aeruginosa PAO1 produces N-oxododecanoyl-l-HSL (3-oxo-C12-HSL) and N-oxooctanoyl-l-HSL (3-oxo-C8-HSL) via the LasI synthase, and N-butanoyl-l-HSL (C4-HSL) [15] and N-hexanoyl-l-HSL (C6-HSL) via the RhlI synthase [16].
Results and discussion
Detection of AHLs in real time using a liquid co-culture assay
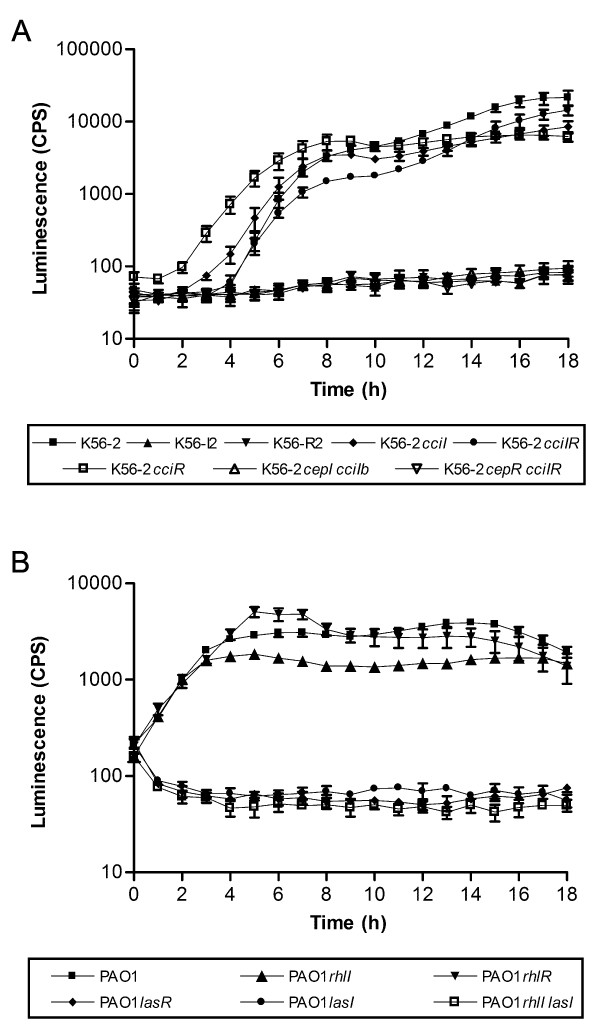
In this study, we examined the utility of the A. tumefaciens A136 (pCF218) (pMV26) reporter system for semi-quantitative detection of AHLs during growth in co-culture with previously characterized QS mutants of B. cenocepacia and P. aeruginosa (Table 1). To monitor the production of AHLs over time, we co-cultured in liquid medium K56-2, PAO1, or their respective QS mutant strains (Table 1) with the A. tumefaciens A136 (pCF218) (pMV26) reporter strain and measured the luminescence over a period of 18 h (Fig. 1A and Fig. 1B). In parallel experiments, strains were grown in the absence of the reporter strain to confirm that the growth rates of the various QS mutants were not affected by the QS gene mutation (data not shown). The AHL expression levels of all B. cenocepacia QS mutant strains (Fig. 1A) are similar to those previously reported using thin-layer chromatography bioassay analysis [12,13]. In B. cenocepacia K56-2 no AHLs were detected when either cepI or cepR was mutated since CepR is essential for the expression of both the CepI/R and CciI/R QS systems [13]. Intermediate amounts of AHL were detected in cciI or cciR mutants. Interestingly, AHL production is detected earlier in growth in the cciR mutant. CciR negatively regulates cepI which likely accounts for the earlier expression of AHLs [13]. In P. aeruginosa, LasR is at the top of the hierarchy of both QS systems [17] and regulates expression of both lasI and rhlI synthases (Fig. 1B). Therefore lasR or lasI mutants have little detectable AHL, whereas the rhlI mutation results in a slight decrease in AHLs in the culture medium (Fig. 1B).
Table 1.
Genotypes of bacterial strains used in this study.
| Strain a | Genotype | Reference or source |
| B. cenocepacia | ||
| K56-2 | Wild type | [21] |
| K56-I2 | cepI | [11] |
| K56-dI2 | cepI | [13] |
| K56-R2 | cepR | [11] |
| K56-2cciI | cciI | [13] |
| K56-2cciIR | cciI, cciR | [13] |
| K56-2cciR | cciR | [13] |
| K56-2cepI cciIb | cepI, cciI | [13] |
| K56-2cepR cciIR | cepR, cciI, cciR | [13] |
| P. aeruginosa | ||
| PAO1 | Wild type | [22] |
| PAO1rhlI (PDO100) | rhlI | [23] |
| PAO1rhlR (PDO111) | rhlR | [23] |
| PAO1lasR (PAO-R1) | lasR | [24] |
| PAO1lasI (PAO214) | lasI | H. P. Schweizer |
| PAO1rhlI lasI (PAO-JP2) | rhlI, lasI | [25] |
a Names in parentheses are previously published strain names.
Figure 1.
Detection of AHL using the A. tumefaciens A136 (pCF218) (pMV26) as biosensor in liquid co-culture assay. Detection and temporal expression of AHL in liquid co-culture assay for B. cenocepacia (A) and P. aeruginosa strains (B). Values shown are the mean ± standard deviation of at least four replicates.
If this assay was employed with test strains with varying growth rates, the luminescence values could be normalized to optical density, although it would need to be noted that the normalized value would be the optical density of the combined cultures and not necessarily reflective of the cell density of the strain being analyzed for AHL production. The only strain shown in Fig. 1 with slower growth was PAO1rhlIlasI which doesn't produce AHLs regardless of growth phase. The strains shown in Fig. 1A and Fig. 1B reached similar OD 600 nm values in the co-culture assay suggesting that there were no growth affects due to co-culture.
We determined that K56-2 did not alter growth of the reporter strain by comparing the number of A136 present in co-culture or single culture at 0 and 24 hr. Cultures were plated on LB agar and B. cepacia selective agar (BCSA) [18]. The number of cfu recovered on BCSA was subtracted from the number of cfu recovered on L agar to determine the number of A136 present. There was no difference in the number of A136 present grown alone or with K56-2 and the ratio of A136 to K56-2 was similar at both timepoints (data not shown).
Detection of AHLs using a soft agar co-culture assay
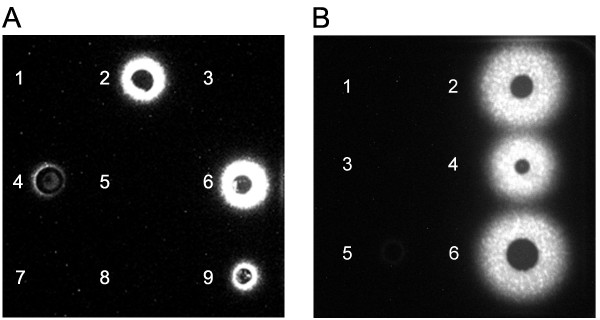
We previously developed a soft agar co-culture assay to detect AHL production in B. cenocepacia [19]. In this study, we determined if this assay could be used to qualitatively assess AHL production and if the results correlated with liquid co-culture assay. In this assay, the size of the AHL diffusion ring within the soft agar is directly proportional to the amount of AHL produced by the bacterial strain. When comparing the B. cenocepacia results on soft agar (Fig. 2A) to the results of the liquid co-culture assay (Fig. 1A), we observed that the non-producing AHL strains were negative in both methods. However, both cciI and cciR mutant strains produced more luminescence than the wild type K56-2 strain unlike in the liquid assay (Fig. 1A). It is possible that growth differences on the soft agar or regulatory pathways affected differently by these mutations on agar surfaces may explain the differences between the two assays for these strains. For P. aeruginosa strains, results from the soft agar assay could be extrapolated to the liquid co-culture assay (Fig. 2B).
Figure 2.
Detection of AHLs using the A. tumefaciens A136 (pCF218) (pMV26) as biosensor on soft agar co-culture assay. Detection of AHLs in a soft agar co-culture assay for B. cenocepacia (A) and P. aeruginosa strains (B). Panel A: 1, K56-2cepI cciIb; 2, K56-2cciI; 3, K56-2cepR cciIR; 4, K56-2cciIR; 5, K56-dI2; 6, K56-2cciR; 7, K56-I2; 8, K56-R2; 9, K56-2. Panel B: 1, PAO1rhlI lasI; 2, PAO1rhlR; 3, PAO1lasI; 4, PAO1rhlI; 5, PAO1lasR; 6, PAO1.
Application of the liquid co-culture assay as a high-throughput screening tool
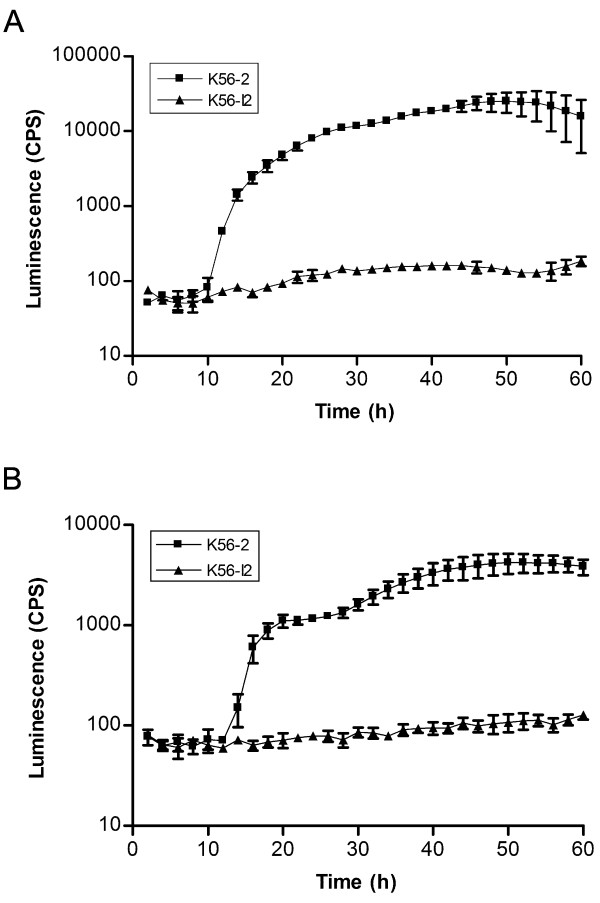
The liquid co-culturing system is adaptable for high-throughput screening. To demonstrate this we evaluated the assay using both a 96- and a 384-well plate format (Fig. 3) with B. cenocepacia strains K56-2 and K56-I2 (cepI) (Table 1). Although the CPS are lower in the 384-well format (Fig. 3B), due to reduced growth (data not shown), trends between the two formats were identical indicating that the 384-well plate format could also be utilized as a semi-quantitative assay.
Figure 3.
Comparison of the detection of AHLs using the A. tumefaciens A136 (pCF218) (pMV26) as biosensor with different microtiter plate formats. Temporal expression of AHL in liquid co-culture assay by B. cenocepacia strains in 96- (A) and 384-well formats (B). Values shown are the mean ± standard deviation of at least 3 replicates.
Conclusion
In summary, the co-culture assays described in both liquid and soft agar enable the detection and semi-quantitation of AHL production in P. aeruginosa and B. cenocepacia (Fig. 1 and Fig. 2) and should be applicable to AHL analysis in other bacterial species, providing the strains could grow in co-culture with the reporter strain. The reporter is sensitive to a wide range of AHLs [6]. However, the sensitivity of the luminescence counter and the camera system used is critical for the successful detection of a broad range of AHLs [6]. These two assays have enormous potential as high-throughput screening tools especially with the use of the 384-well format. Large libraries can quickly be screened and mutants defective in producing AHL easily identified by measuring CPS with the liquid assay. However, one of the most promising applications for this AHL detection assay might be for the identification of QS blockers by screening compound libraries in 384-well formats and looking for the absence of signal. Potential mutants or QS blockers identified using this screening assay could be validated using subsequent assays for the detection and measurement of specific AHLs, as well as assays that correlated AHL production to growth. With the increasing numbers of bacterial species resistant to all or most available antibiotics, alternative therapeutic approaches are needed. One strategy is to target bacterial virulence mechanisms [20]. Since QS systems control the expression of several virulence factors in many pathogens, compounds that block production of AHL signals or interfere with their activity may be effective anti-virulence agents.
Methods
Bacterial strains, plasmids and growth conditions
A. tumefaciens A136 (pCF218) (pMV26) [5,6] was grown in Luria-Bertani broth (LB) (Miller's broth base, Invitrogen, Burlington, Ontario, Canada) at 28 – 30°C for up to 24 h with the addition of 25 μg/ml of kanamycin and 4.5 μg/ml of tetracycline. B. cenocepacia and P. aeruginosa strains were grown in Trypticase soy broth (TSB; Becton, Dickinson and Company, Sparks, MD, USA) and LB respectively with antibiotics if required and incubated at 37°C overnight. Chemicals were purchased from Sigma-Aldrich Canada, Ltd., (Oakville, Ontario, Canada).
Luminescence bioassays
For the detection and monitoring of AHL production, bioluminescence assays employing soft agar plates or liquid medium were developed using A. tumefaciens A136 (pCF218) (pMV26) as a reporter strain [5,6]. The soft agar co-culture assay was performed as previously described [19]. Briefly, an overnight culture of the reporter strain was mixed in a ratio 1:80 (v/v) with a mixture of TSB (Becton, Dickinson and Company) containing 0.7% agar (w/v). Each plate was prepared with 20 ml of the TSB agar reporter strain mixture and let dry for approximately 2 h at room temperature before use. Two μl of an overnight culture normalized to an OD600 of 0.3 of B. cenocepacia or P. aeruginosa were spotted onto the soft agar of the reporter plate, let dry for 20 min, and incubated at 28 – 30°C for 24 h. Pictures of the reporter plates were taken using a Fluorchem™ 8900 digital camera system (Alpha Innotech, San Leandro, California, USA) to detect luminescence. For the liquid co-culture assay, 150 μl or 75 μl of TSB (Becton, Dickinson and Company) or LB (Invitrogen) were added to each well of black, clear-bottom 96- and 384-well microtiter plates (Corning Inc., Corning, NY, USA), respectively. An overnight culture of the A. tumefaciens A136 (pCF218) (pMV26) reporter strain previously diluted 1:10 (v/v) was added to each well for a final dilution of 1:1500 (v/v). One μl of an overnight culture of B. cenocepacia K56-2 was added to each well containing the reporter strain. To prevent evaporation, 75 and 25 μl of mineral oil were added on top of each well of the 96- and 384-well plates, respectively. Plates were placed into a Wallac Victor2 Model 1420 Multi-label Counter (Perkin-Elmer Life Sciences, Boston, MA, USA), incubated at 28 – 30°C with constant shaking and luminescence was detected (CPS, counts per second) at hourly intervals. At least three replicates were performed for each assay.
Authors' contributions
SPB developed the soft agar and the liquid co-culture assays, and drafted the manuscript. ALB helped develop the liquid co-culture assays. PAS supervised the study and contributed to the drafting of the manuscript. All authors have approved the final version of the manuscript.
Acknowledgments
Acknowledgements
This study was supported by the Canadian Cystic Fibrosis Foundation (CCFF) special initiative in memory of Michael O'Reilly. SB was the recipient of a studentship from CCFF. The authors thank DF Viteri for excellent technical assistance.
Contributor Information
Steve P Bernier, Email: sbernier@pasteur.fr.
Anne L Beeston, Email: beestona@inspection.gc.ca.
Pamela A Sokol, Email: psokol@ucalgary.ca.
References
- Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht J, Silverman M. Identification of genes and gene products necessary for bacterial bioluminescence. Proc Natl Acad Sci U S A. 1984;81:4154–4158. doi: 10.1073/pnas.81.13.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead NA, Barnard AM, Slater H, Simpson NJ, Salmond GP. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol Rev. 2001;25:365–404. doi: 10.1111/j.1574-6976.2001.tb00583.x. [DOI] [PubMed] [Google Scholar]
- Steindler L, Venturi V. Detection of quorum-sensing N-acyl homoserine lactone signal molecules by bacterial biosensors. FEMS Microbiol Lett. 2007;266:1–9. doi: 10.1111/j.1574-6968.2006.00501.x. [DOI] [PubMed] [Google Scholar]
- Sokol PA, Sajjan U, Visser MB, Gingues S, Forstner J, Kooi C. The CepIR quorum-sensing system contributes to the virulence of Burkholderia cenocepacia respiratory infections. Microbiology. 2003;149:3649–3658. doi: 10.1099/mic.0.26540-0. [DOI] [PubMed] [Google Scholar]
- Chambers CE, Visser MB, Schwab U, Sokol PA. Identification of N-acylhomoserine lactones in mucopurulent respiratory secretions from cystic fibrosis patients. FEMS Microbiol Lett. 2005;244:297–304. doi: 10.1016/j.femsle.2005.01.055. [DOI] [PubMed] [Google Scholar]
- Watson B, Currier TC, Gordon MP, Chilton MD, Nester EW. Plasmid required for virulence of Agrobacterium tumefaciens. J Bacteriol. 1975;123:255–264. doi: 10.1128/jb.123.1.255-264.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Beaber JW, More MI, Fuqua C, Eberhard A, Winans SC. Analogs of the autoinducer 3-oxooctanoyl-homoserine lactone strongly inhibit activity of the TraR protein of Agrobacterium tumefaciens. J Bacteriol. 1998;180:5398–5405. doi: 10.1128/jb.180.20.5398-5405.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meighen EA, Szittner RB. Multiple repetitive elements and organization of the lux operons of luminescent terrestrial bacteria. J Bacteriol. 1992;174:5371–5381. doi: 10.1128/jb.174.16.5371-5381.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol PA, Malott RJ, Riedel K, Eberl L. Communication systems in the genus Burkholderia: global regulators and targets for novel antipathogenic drugs. Future Microbiol. 2007;2:555–563. doi: 10.2217/17460913.2.5.555. [DOI] [PubMed] [Google Scholar]
- Lewenza S, Conway B, Greenberg EP, Sokol PA. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J Bacteriol. 1999;181:748–756. doi: 10.1128/jb.181.3.748-756.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewenza S, Sokol PA. Regulation of ornibactin biosynthesis and N-acyl-L-homoserine lactone production by CepR in Burkholderia cepacia. J Bacteriol. 2001;183:2212–2218. doi: 10.1128/JB.183.7.2212-2218.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malott RJ, Baldwin A, Mahenthiralingam E, Sokol PA. Characterization of the cciIR quorum-sensing system in Burkholderia cenocepacia. Infect Immun. 2005;73:4982–4992. doi: 10.1128/IAI.73.8.4982-4992.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M, Eberl L, Tummler B. Quorum sensing: the power of cooperation in the world of Pseudomonas. Environ Microbiol. 2005;7:459–471. doi: 10.1111/j.1462-2920.2005.00769.x. [DOI] [PubMed] [Google Scholar]
- Pearson JP, Gray KM, Passador L, Tucker KD, Eberhard A, Iglewski BH, Greenberg EP. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci U S A. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winson MK, Camara M, Latifi A, Foglino M, Chhabra SR, Daykin M, Bally M, Chapon V, Salmond GP, Bycroft BW, Lazdunski A, Stewart GSAB, Williams P. Multiple N-acyl-L-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1995;92:9427–9431. doi: 10.1073/pnas.92.20.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesci EC, Pearson JP, Seed PC, Iglewski BH. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry DA, Campbell ME, LiPuma JJ, Speert DP. Identification of Burkholderia cepacia isolates from patients with cystic fibrosis and use of a simple new selective medium. J Clin Microbiol. 1997;35:614–619. doi: 10.1128/jcm.35.3.614-619.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier SP, Nguyen DT, Sokol PA. A LysR-type transcriptional regulator in Burkholderia cenocepacia influences colony morphology and virulence. Infect Immun. 2008;76:38–47. doi: 10.1128/IAI.00874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ. The biology and future prospects of antivirulence therapies. Nature reviews. 2008;6:17–27. doi: 10.1038/nrmicro1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahenthiralingam E, Coenye T, Chung JW, Speert DP, Govan JR, Taylor P, Vandamme P. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J Clin Microbiol. 2000;38:910–913. doi: 10.1128/jcm.38.2.910-913.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway BW, Romling U, Tummler B. Genomic mapping of Pseudomonas aeruginosa PAO. Microbiology. 1994;140 ( Pt 11):2907–2929. doi: 10.1099/13500872-140-11-2907. [DOI] [PubMed] [Google Scholar]
- Brint JM, Ohman DE. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J Bacteriol. 1995;177:7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambello MJ, Iglewski BH. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JP, Pesci EC, Iglewski BH. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]