Abstract
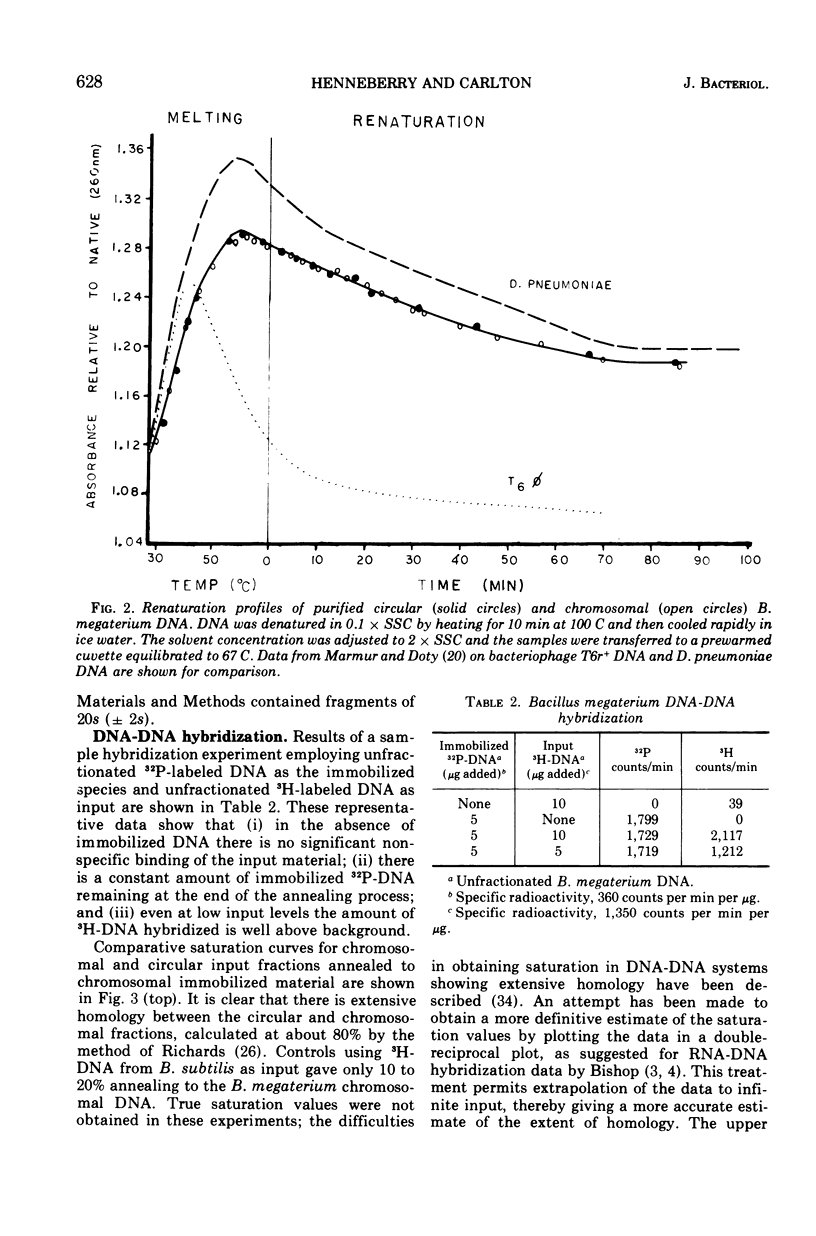
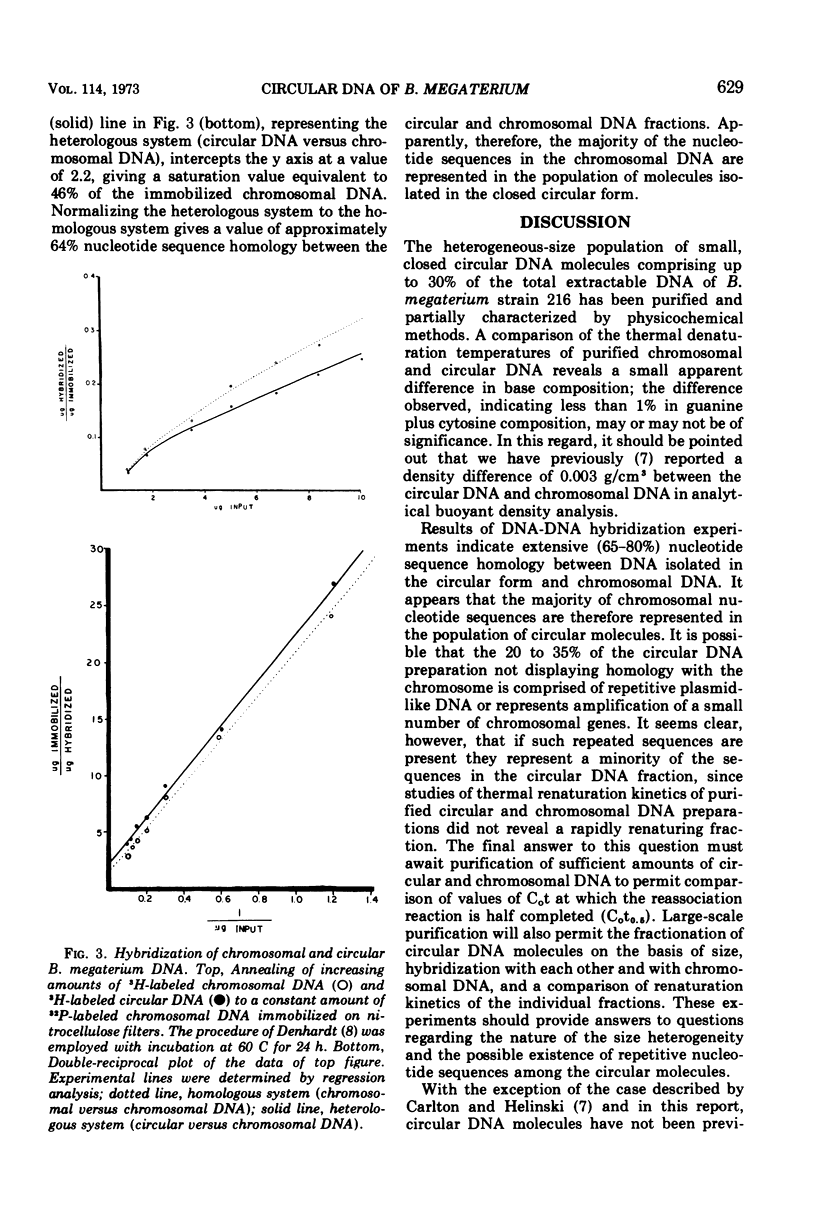
The polydisperse circular deoxyribonucleic acid (DNA) molecules which comprise up to 30% of the total extractable DNA of Bacillus megaterium strain 216 have been purified and partially characterized. Banding in cesium chlorideethidium bromide by “gradient relaxation” in a fixed-angle rotor provided good resolution of circular and chromosomal DNAs for preparative separations. Renaturation studies on purified circular DNA failed to reveal a rapidly renaturing fraction, and DNA-DNA hybridization studies indicated that the majority of the chromosomal nucleotide sequences are represented in the heterogeneous-size population of circular molecules. It is concluded that the circular DNA of B. megaterium does not represent typical bacterial plasmid DNA. The possibility that the circular DNA molecules are the result of the expression of a defective bacteriophage is discussed.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anet R., Strayer D. R. Density gradient relaxation: a method for preparative buoyant density separation of DNA. Biochem Biophys Res Commun. 1969 Feb 7;34(3):328–334. doi: 10.1016/0006-291x(69)90836-5. [DOI] [PubMed] [Google Scholar]
- BURTON K. A study of the conditions and mechanism of the diphenylamine reaction for the colorimetric estimation of deoxyribonucleic acid. Biochem J. 1956 Feb;62(2):315–323. doi: 10.1042/bj0620315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazaral M., Helinski D. R. Characterization of multiple circular DNA forms of colicinogenic factor E-1 from Proteus mirabilis. Biochemistry. 1968 Oct;7(10):3513–3520. doi: 10.1021/bi00850a028. [DOI] [PubMed] [Google Scholar]
- Bishop J. O. Interpretation of DNA-RNA hybridization data. Nature. 1969 Nov 8;224(5219):600–603. doi: 10.1038/224600a0. [DOI] [PubMed] [Google Scholar]
- Bishop J. O., Robertson F. W. Transcription of bacteriophage T4 deoxyribonucleic acid in vitro. Biochem J. 1969 Nov;115(3):353–361. doi: 10.1042/bj1150353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton B. C., Helinski D. R. Heterogeneous circular DNA elements in vegetative cultures of Bacillus megaterium. Proc Natl Acad Sci U S A. 1969 Oct;64(2):592–599. doi: 10.1073/pnas.64.2.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Haas M., Yoshikawa H. Defective bacteriophage PBSH in Bacillus subtilis. I. Induction, purification, and physical properties of the bacteriophage and its deoxyribonucleic acid. J Virol. 1969 Feb;3(2):233–247. doi: 10.1128/jvi.3.2.233-247.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas M., Yoshikawa H. Defective bacteriophage PBSH in Bacillus subtilis. II. Intracellular development of the induced prophage. J Virol. 1969 Feb;3(2):248–260. doi: 10.1128/jvi.3.2.248-260.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helinski D. R., Clewell D. B. Circular DNA. Annu Rev Biochem. 1971;40:899–942. doi: 10.1146/annurev.bi.40.070171.004343. [DOI] [PubMed] [Google Scholar]
- IVANOVICS G., ALFOLDI L., LOVAS B. Cultivation and electron microscopy of a bacteriocinogenic strain of Bacillus megaterium. Acta Microbiol Acad Sci Hung. 1957;4(3):295–308. [PubMed] [Google Scholar]
- Johnson J. L., Ordal E. J. Deoxyribonucleic acid homology in bacterial taxonomy: effect of incubation temperature on reaction specificity. J Bacteriol. 1968 Mar;95(3):893–900. doi: 10.1128/jb.95.3.893-900.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KECK K. An ultramicro technique for the determination of deoxypentose nucleic acid. Arch Biochem Biophys. 1956 Aug;63(2):446–451. doi: 10.1016/0003-9861(56)90059-5. [DOI] [PubMed] [Google Scholar]
- Leavitt R. W., Wohlhieter J. A., Johnson E. M., Olson G. E., Baron L. S. Isolation of circular deoxyribonucleic acid from Salmonella typhosa hybrids obtained from matings with Escherichia coli Hfr donors. J Bacteriol. 1971 Dec;108(3):1357–1365. doi: 10.1128/jb.108.3.1357-1365.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol. 1962 Jul;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- MARMUR J., DOTY P. Thermal renaturation of deoxyribonucleic acids. J Mol Biol. 1961 Oct;3:585–594. doi: 10.1016/s0022-2836(61)80023-5. [DOI] [PubMed] [Google Scholar]
- Novick R. P. Extrachromosomal inheritance in bacteria. Bacteriol Rev. 1969 Jun;33(2):210–263. doi: 10.1128/br.33.2.210-263.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K., Mudd J. A., Mangan J., Huang W. M., Subbaiah T. V., Marmur J. Properties of the defective phage of Bacillus subtilis. J Mol Biol. 1968 Jun 28;34(3):413–428. doi: 10.1016/0022-2836(68)90169-1. [DOI] [PubMed] [Google Scholar]
- Okamoto K., Mudd J. A., Marmur J. Conversion of Bacillus subtilis DNA to phage DNA following mitomycin C induction. J Mol Biol. 1968 Jun 28;34(3):429–437. doi: 10.1016/0022-2836(68)90170-8. [DOI] [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards O. C. Hybridization of Euglena gracilis chloroplast and nuclear DNA. Proc Natl Acad Sci U S A. 1967 Jan;57(1):156–163. doi: 10.1073/pnas.57.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogolsky M., Slepecky R. A. Elimination of a genetic determinant for sporulation of Bacillus subtilis with acriflavin. Biochem Biophys Res Commun. 1964 Jun 15;16(3):204–208. doi: 10.1016/0006-291x(64)90326-2. [DOI] [PubMed] [Google Scholar]
- Roth T. F., Hayashi M. Allomorphic forms of bacteriophage phiX-174 replicative DNA. Science. 1966 Nov 4;154(3749):658–660. doi: 10.1126/science.154.3749.658. [DOI] [PubMed] [Google Scholar]
- SEAMAN E., TARMY E., MARMUR J. INDUCIBLE PHAGES OF BACILLUS SUBTILIS. Biochemistry. 1964 May;3:607–613. doi: 10.1021/bi00893a001. [DOI] [PubMed] [Google Scholar]
- Scaife J. Episomes. Annu Rev Microbiol. 1967;21:601–638. doi: 10.1146/annurev.mi.21.100167.003125. [DOI] [PubMed] [Google Scholar]
- Spizizen J. TRANSFORMATION OF BIOCHEMICALLY DEFICIENT STRAINS OF BACILLUS SUBTILIS BY DEOXYRIBONUCLEATE. Proc Natl Acad Sci U S A. 1958 Oct 15;44(10):1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinograd J., Lebowitz J., Watson R. Early and late helix-coil transitions in closed circular DNA. The number of superhelical turns in polyoma DNA. J Mol Biol. 1968 Apr 14;33(1):173–197. doi: 10.1016/0022-2836(68)90287-8. [DOI] [PubMed] [Google Scholar]
- Warmaar S. O., Cohen J. A. A quantitative assay for DNA-DNA hybrids using membrane filters. Biochem Biophys Res Commun. 1966 Aug 23;24(4):554–558. doi: 10.1016/0006-291x(66)90356-1. [DOI] [PubMed] [Google Scholar]
- Wetmur J. G., Davidson N. Kinetics of renaturation of DNA. J Mol Biol. 1968 Feb 14;31(3):349–370. doi: 10.1016/0022-2836(68)90414-2. [DOI] [PubMed] [Google Scholar]


