Figure 5.
XopD Has Two EAR Motifs Required for Virulence in Tomato and Tissue Necrosis in N. benthamiana.
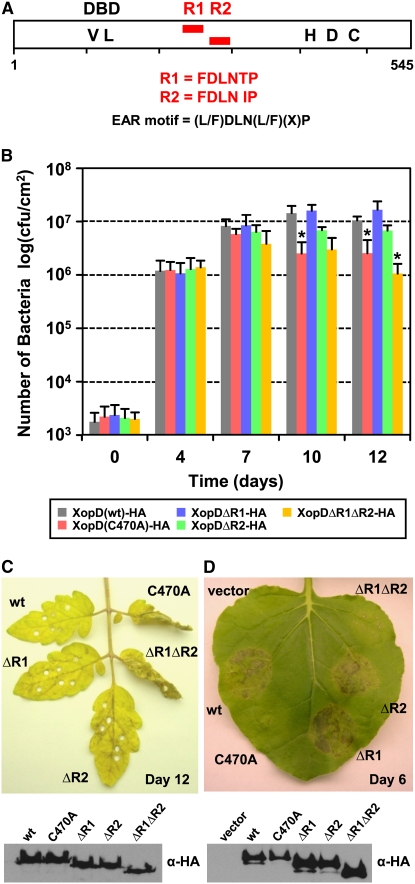
(A) Schematic view of XopD protein. The open rectangle represents XopD protein, and two EAR motifs, R1 and R2, are represented by red rectangles. The motifs are located between the N-terminal DBD and C-terminal SUMO protease domain containing catalytic core residues His, Asp, and Cys (HDC).
(B) XopD EAR motifs are required for maximal Xcv growth in tomato. Susceptible VF36 tomato leaves were hand-inoculated with a 1 × 105 cfu/mL suspension of Xcv ΔxopD expressing XopD(wt)-HA (gray bars), XopD(C470A)-HA (red bars), XopDΔR1-HA (blue bars), XopDΔR2-HA (green bars), or XopDΔR1ΔR2-HA (yellow bars). Data points represent mean log10 cfu per cm2 ± sd of three independent experiments. The asterisk above the bars indicates statistically significant (t test, P < 0.05) differences between the bacterial numbers of Xcv ΔxopD expressing XopD(wt)-HA and the XopD mutant proteins.
(C) XopD EAR motifs are required to suppress disease symptoms in tomato (top panel). Representative phenotype of VF36 tomato leaves inoculated in (B) after 12 d: wt, XopD(wt)-HA; C470A, XopD(C470A)-HA; ΔR1, XopDΔR1-HA; ΔR2, XopDΔR2-HA; ΔR1ΔR2, XopDΔR1ΔR2-HA. Immunoblot analysis (bottom panel) of proteins expressed in the bacteria. Xcv strains described in (B) were grown in XVM2 medium for 6 h with appropriate antibiotics. Total cellular lysate (∼5 × 107 cells) was examined by protein gel blot analysis using HA antisera.
(D) XopDΔR1ΔR2-HA does not elicit necrosis in N. benthamiana (top panel). Leaf was infiltrated with the A. tumefaciens strains expressing the following: vector, vector control; wt, XopD(wt)-HA; C470A, XopD(C470A)-HA; ΔR1, XopDΔR1-HA; ΔR2, XopDΔR2-HA; ΔR1ΔR2, XopDΔR1ΔR2-HA. The phenotype was photographed 6 DAI. Immunoblot analysis (bottom panel) of proteins transiently expressed in N. benthamiana leaves as shown in top panel. Total protein (∼50μg) was extracted from tissue at 48 h after inoculation and then examined by protein gel blot analysis using HA antisera. These experiments were repeated three times with similar results.