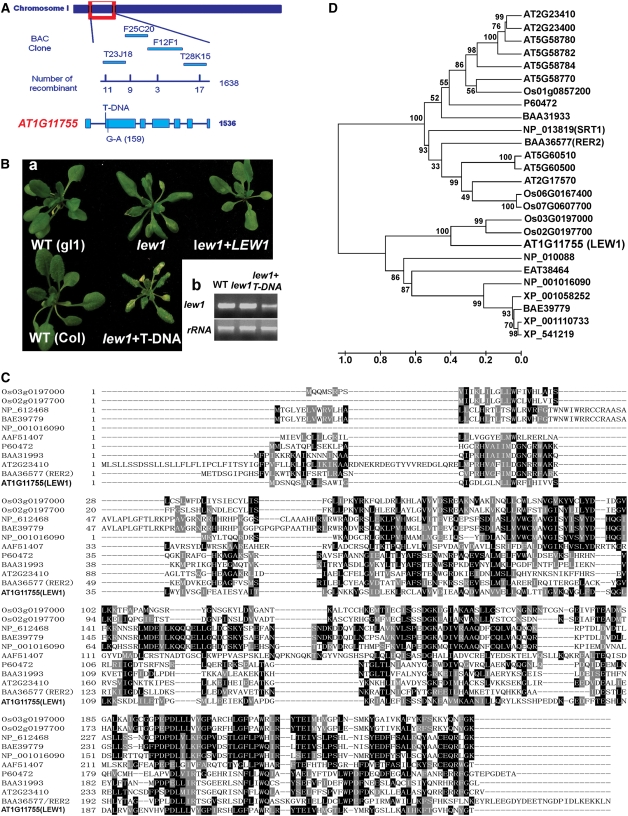
Figure 3.
Position Cloning of LEW1 and Sequence Analysis of Long-Chain cis-Prenyltransferases.
(A) Positional cloning of LEW1. The LEW1 locus was mapped to between two polymorphic markers on BAC clones T23J18 (recombinants, 11 of 1638) and T28K15 (recombinants, 17 of 1638). Further mapping delimited the LEW1 locus to a region within BACs F12F1 (recombinants, 3 of 1638) and T28K15. All candidate open reading frames were sequenced in this region, and only one mutation (G159A) was found in AT1G11755.
(B) (a) Complementation of the lew1 mutant. Shown are the wild type (gl1), the lew1 mutant, the lew1 mutant transformed with 35S-LEW1 (cDNA), the wild type (Columbia [Col]), and a heterozygous plant of lew1 crossed with a line in which the LEW1 gene was disrupted by a T-DNA insertion. The T-DNA was inserted at position 205 counting from the first putative ATG of AT1G11755. (b) RT-PCR analysis of LEW1 transcripts in the wild type, lew1, and a heterozygous plant of lew1 crossed with the T-DNA line. rRNA is shown as a control for loading.
(C) Alignment of long-chain cis-prenyltransferases from different organisms: rice, Os02g0197700 and Os03g0197000; human, NP_612468; mouse, BAE39779; Xenopus, NP_001016090; fruit fly, AAF51407; E. coli, P60472; Micrococcus luteus, BAA31993; Saccharomyces cerevisiae, BAA36577 (RER2); Arabidopsis, AT2G23410 and AT1G11755 (LEW1).
(D) Phylogenetic tree of LEW1 and related proteins from different organisms: Arabidopsis, At2g23400, At5g58780, At5g58782, At5g58784, At5g58770, At5g60510, At5g60500, and At2g17570; rice, Os01g0857200, Os06g0167400, and Os07g0607700; monkey, XP_001110733; dog, XP_541219; Aedes aegypti, EAT38464; rat, XP_001058252; S. cerevisiae, NP_013819/SRT1 and NP_010088; the others are the same as in (C). The phylogenetic tree was constructed on the alignment using MEGA (version 4.0) (see Supplemental Data Set 1 online). Bootstrap values were calculated from 1000 trials and are shown at each node. The extent of divergence according to the scale (relative units) is shown at bottom.