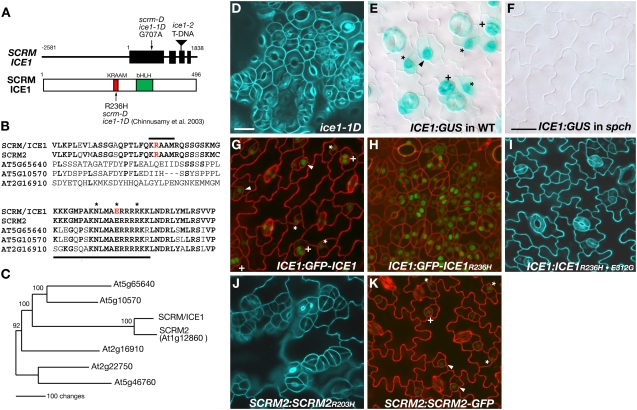
Figure 4.
SCRM Is ICE1.
(A) SCRM/ICE1 gene (top) and protein (bottom). Top: line, noncoding sequence; box, exon; triangle, T-DNA insertion. +1 corresponds to A at the initiation site (ATG). Bottom: 1 corresponds to the first Met at the initiation codon; red, KRAAM motif; green, bHLH domain. The locations of the mutation are indicated.
(B) Amino acid alignment of ICE1 and related bHLH proteins. Top: the KRAAM motif (marked by a solid line); red, the Arg residue substituted by His in scrm-D, ice1-1D, and SCRM2R203H. Bottom: the basic region (underlined); red, the invariant Glu required for DNA binding; asterisks, core amino acid residues for DNA binding.
(C) Molecular phylogeny of ICE1 and related bHLHs. Shown is a parsimony tree based on the entire coding sequence (see alignment in Supplemental Data Set 1 online). Branch length reflects the number of amino acid changes. Bootstrap values (%) for 1000 replications are indicated on each node.
(D) Abaxial cotyledon epidermis of ice1-1D.
(E) ICE1:GUS expression (blue) in wild-type abaxial cotyledon epidermis.
(F) ICE1:GUS expression (blue) in spch abaxial cotyledon epidermis.
(G) Localization of GFP-ICE1 fusion protein (green) expressed by the native ICE1 promoter.
(H) Localization of GFP-ICE1R236H fusion protein (green) expressed by the native ICE1 promoter.
(I) Abaxial rosette leaf epidermis of ICE1:ICE1R236H·E312G.
(J) Abaxial rosette leaf epidermis of SCRM2:SCRM2R203H rosette leaves.
(K) Localization of SCRM2-GFP fusion protein (green) expressed by the native SCRM2 promoter.
For (E), (G), and (K), meristemoids are indicated by asterisks, GMCs by arrowheads, and immature guard cells by plus signs. Bars = 20 μm.