Abstract
Peptide nucleic acid (PNA) oligomers targeted to guanine quadruplex-forming RNAs can be designed in two different ways. First, complementary cytosine-rich PNAs can hybridize by formation of Watson-Crick base pairs, resulting in hybrid PNA-RNA duplexes. Second, guanine-rich homologous PNAs can hybridize by formation of G-tetrads, resulting in hybrid PNA-RNA quadruplexes. UV thermal denaturation, circular dichroism and fluorescence spectroscopy experiments were used to compare these two recognition modes and revealed 1:1 duplex formation for the complementary PNA and 2:1 (PNA2-RNA) quadruplex formation for the homologous PNA. Both hybrids were very stable and hybridization was observed at low nanomolar concentrations. Hybrid quadruplex formation was equally efficient regardless of the PNA strand polarity, indicating a lack of interaction between the loop nucleobases on the PNA and RNA strands. The implications of this finding on sequence specificity as well as methods to improve affinity are also discussed.
The guanine (G) quadruplex structure arises from the simultaneous hydrogen bonding of four guanine nucleobases in a quasi-planar array, with multiple such G-quartets associated cofacially via pi stacking (1–3). Binding of specific metal ions within the G-tetrad stack stabilizes the overall structure (4,5). G quadruplexes are formed by oligonucleotides based on telomeric repeat sequences (5) and have been implicated in several biological processes (6), including regulating transcription (7–9), mediating RNA-protein interactions (10–14), and controlling alternative RNA splicing (15,16) and translation (17). They are also commonly selected from combinatorial synthetic DNA or RNA libraries screened for binding to a variety of protein targets (12,18–23).
The growing awareness of the biological importance of G quadruplexes has motivated considerable interest in developing synthetic compounds that can bind selectively to these structures (24). A wide variety of G quadruplex-binding ligands have been discovered and these exhibit varying degrees of affinity and selectivity, with selectivity relating both to quadruplex versus duplex nucleic acid structures as well as between different quadruplex topologies (25,26). Thus, these compounds are intended to recognize specific nucleic acid structures.
Our research focuses on targeting G quadruplex-forming sequences, rather than G quadruplex structures. In one approach, a Watson-Crick complementary oligomer is synthesized and targeted to some sub-sequence within the overall G quadruplex target. Because of the high percentage of guanine in the target, a large number of G-C pairs are formed upon hybridization of the complementary oligomer. Combining a high G-C content with a high affinity DNA mimic such as peptide nucleic acid (PNA) allows very short complementary oligomers to bind and form stable hybrid duplexes. This strategy has been shown to be effective for both DNA (27,28) and RNA (29) quadruplex targets.
An alternative approach to recognition of quadruplex forming sequences involves synthesizing homologous oligomers, in which case both the target and the probe have the same G-rich sequence of nucleobases. Homologous PNAs form hybrid G quadruplexes with both DNA (30) and RNA targets (29). These structures exhibit stabilization by specific metal ions such as potassium and sodium (but not lithium), analogous to pure DNA and RNA quadruplexes. Moreover, the hybrid quadruplexes are very stable and readily form at low nanomolar concentrations (or even lower). Thus, a sequence-targeted approach to recognizing these important regulatory regions of DNA or RNA has the potential to create high affinity, specific regulators of gene expression.
We recently compared complementary and homologous hybridization of PNA probes to an RNA quadruplex and found that 7 base PNAs were able to disrupt the folded RNA structure in order to bind to their target sequence (29). While the complementary PNA formed the expected 1:1 duplex structure with the RNA target, the homologous PNA surprisingly formed a 2:1 (i.e. PNA2-RNA) hybrid quadruplex. This report describes using the same probes to target a simplified RNA quadruplex, allowing thermodynamic data to be obtained for the hybridization reactions. In addition, homologous PNA probes were synthesized with both possible strand polarities (N-C versus C-N) and used to explore the issue of PNA strand alignment within hybrid quadruplexes. These experiments provide an increasingly detailed picture of hybrid PNA-RNA quadruplex formation and point the way toward further increasing the affinity of short G-rich PNAs for homologous targets in biological RNA.
EXPERIMENTAL PROCEDURES
Chemicals
2’-O-orthoester protected RNA oligonucleotides were purchased from Dharmacon Research (www.dharmacon.com). The RNA oligonucleotides were deprotected using TEMED-acetate buffer (pH = 3.8) according to guidelines provided by the manufacturer. Stock solutions of the RNA oligonucleotide were prepared in water. Concentration determinations were based on the absorption at 260 nm and were measured at 80 ºC on a Cary 3 Bio spectrophotometer. At 80 ºC the nucleobases are assumed to be unstacked and the molar absorptivity is then the sum of individual absorptivities of the RNA monomers. Values for the extinction coefficients for RNA were obtained from literature (31).
t-Boc-protected PNA monomers were purchased from Applied Biosystems (www.appliedbiosystems.com) and were used to synthesize PNA probes by a solid phase peptide synthesis method (32,33). A PNA probe was labeled at the N terminus with a carboxylic acid derivative of the thiazole orange (TO) dye prior to cleavage, following the same procedure as for the coupling of PNA monomers. The synthesis of the thiazole orange derivative is described elsewhere (34,35). PNA probes were purified via HPLC and characterized by mass spectrometry. Mass spectra were recorded on a matrix-assisted laser desorption ionization time-of-flight instrument (MALDI-TOF) (Applied Biosystems Voyager DE sSTR) using α-cyano-4-hydroxycinnamic acid as the matrix (P7C: m/z calculated for [M+H]+ 1970.9, found 1969.5; P7CTO: m/z calculated 2357.9, found 2355.6; P7H: m/z calculated 2121.98, found 2119.78; P7HTO: m/z calculated 2508.28, found 2505.50; P7Hr: m/z calculated 2121.98, found 2120.68). Extinction coefficients for PNA monomers were obtained from Applied Biosystems (C = 6,600 M−1 cm−1 ; T = 8,600 M−1 cm−1; A = 13,700 M−1 cm−1; G = 11,700 M−1 cm−1). All experiments were carried out in buffer containing 10mM TrisHCl (pH = 7.0) and various amounts of KCl or LiCl as denoted in text or figure captions.
UV Melting Curves
UV-Vis measurements were performed on a Varian Cary 3 spectrophotometer equipped with a thermoelectrically controlled multicell holder. RNA-PNA hybrids were prepared by mixing equimolar amounts of the single strands in buffer, heating to 95 ºC, equilibrating for 5 minutes and then cooling (annealing) to 15 ºC at a rate of 1 ºC/min while monitoring the change in absorbance signal at 275 and 295 nm with a data interval of 0.5 ºC. Subsequent reheating under the same condition revealed no hysteresis effect.
Circular Dichroism
CD experiments were performed on a Jasco J-715 spectrophotometer. All spectra represent an average of six scans and were collected at a rate of 100 nm/min. All spectra were baseline subtracted and smoothed via a nine point adjacent averaging algorithm. Hybrids were prepared by heating to 95 ºC followed by slow cooling to 20 ºC prior to recording spectra. Temperature dependent spectra were obtained by annealing to the lowest temperature of interest. The samples were equilibrated for 10 minutes at that temperature, and then scanned (as above). Samples were then heated by 10 ºC at a rate of 1 ºC/min and equilibrated at that temperature for 5 minutes before subsequent scanning.
Fluorescence Spectroscopy
Fluorescence emission spectra were collected on a Photon Technology International (PTI) fluorescence system. The emission spectra of RNA+PNATO samples were measured upon excitation of TO at 495 nm. The spectral band widths used were either 4 nm or 5 nm. Samples containing equimolar amounts of PNA and RNA were prepared in buffer containing 10 mM TrisHCl (pH = 7.0) and 100 mM KCl and annealed prior to recording the spectra.
Determination of Binding Constants
10 μM stock solutions of RNA targets were prepared in buffer containing 10 mM TrisHCl (pH = 7.0) and 100 mM KCl and annealed. RNA targets were titrated into 0.1 μM solutions containing labeled PNAs (either P7HTO or P7CTO) prepared in the same buffer. Fluorescence spectra recorded 10 minutes after each addition of RNA exhibited increasing intensity, indicative of the hybridization of PNAs to the RNA targets. Based on the concentration dependence of the fluorescence and assuming 100% binding at the saturation of fluorescence signal, the fitting to a 1:1 binding model (equation (1)) yielded the equilibrium binding constants (36).
| (1) |
[P]0 represents the concentration of PNA used, [G]0 is the concentration of RNA at each point in the titration and F0, F and F∞ are the fluorescence intensities of: PNA in solution, PNA+RNA at each point of the titration and final value of fluorescence at saturation point, respectively. The “fraction bound” was calculated by dividing the fluorescence at each point in the titration (F) by the fluorescence at the saturation point (F∞ ). The binding constant was calculated as 1/KD.
Determination of Binding Stoichiometry
Continuous variation experiments (37) for the RNA-PNATO binding were performed by mixing variable ratios of TO-labeled PNA and RNA targets together but keeping the total concentration constant (0.5 μM). Samples were prepared in buffer containing 10 mM TrisHCl (pH = 7.0) and 100 mM KCl and were annealed prior to recording fluorescence spectra as described before. Emission at 535 nm was plotted against the mole fraction of PNATO and stoichiometry was determined from the maximum point in the plot.
RESULTS
RNA Target Characterization
RDQ was originally selected from a combinatorial RNA library for binding to the fragile X mental retardation protein (12). While it is not a natural target, it represented a useful RNA quadruplex model system. In our previous report, we targeted RDQ with complementary and homologous PNAs P7C and P7H (Chart 1). The RNA consists of duplex and quadruplex domains based on secondary structure prediction and 1H-NMR experiments (14). The presence of the stem region in the RNA complicates analysis of PNA hybridization due to its background absorbance. Therefore, we designed a truncated version in which the stem is deleted (RQ). The extra nucleotides separating the duplex and quadruplex domains in RDQ were retained in RQ due to the fact that overhanging nucleotides have been shown to stabilize PNA-DNA (27) and PNA-RNA (38) hybrids.
Chart 1.
RNA targets, PNA probes and thiazole orange dye structure. Target sequence for PNAs is shown in blue.
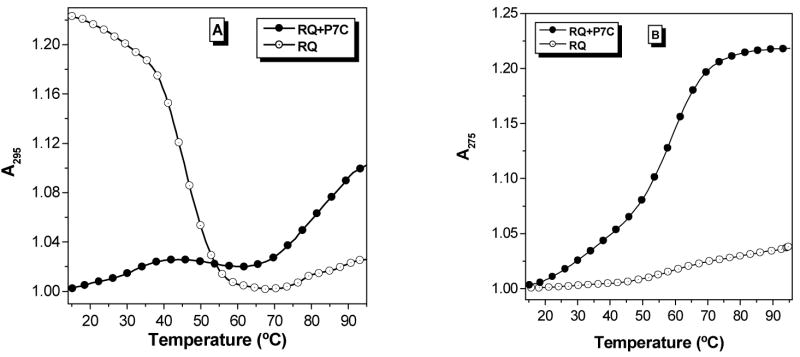
Figure 1 shows CD spectra recorded for the two RNAs in aqueous buffer. The spectra have similar shapes in the region 225–300 nm, but differ at 210 nm. The negative peak evident in the spectrum for RDQ is characteristic of duplex RNA (39), so it is not surprising that this feature is lost when the stem is deleted to give RQ.
Figure 1.
Circular dichroism spectra recorded at 20 °C for RDQ and RQ. [RNA] = 2.0 μM, [KCl] = 100 mM.
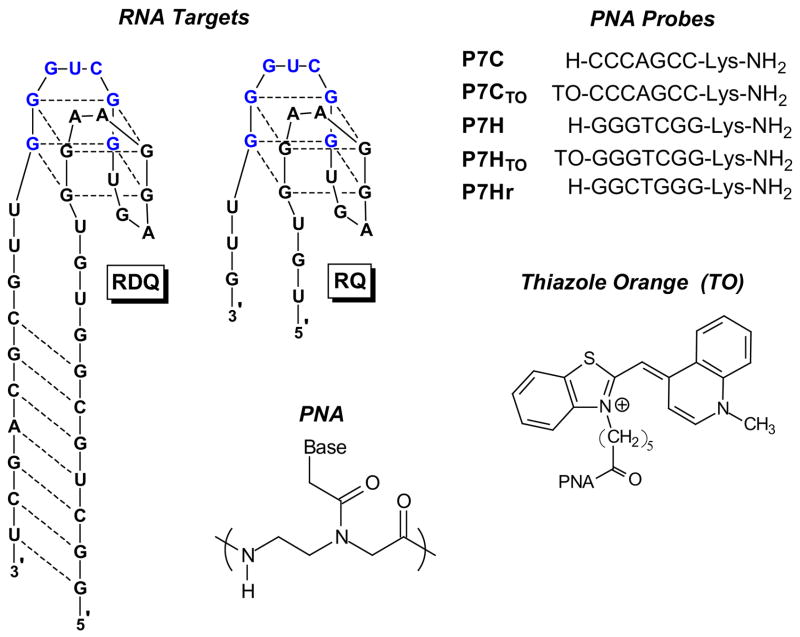
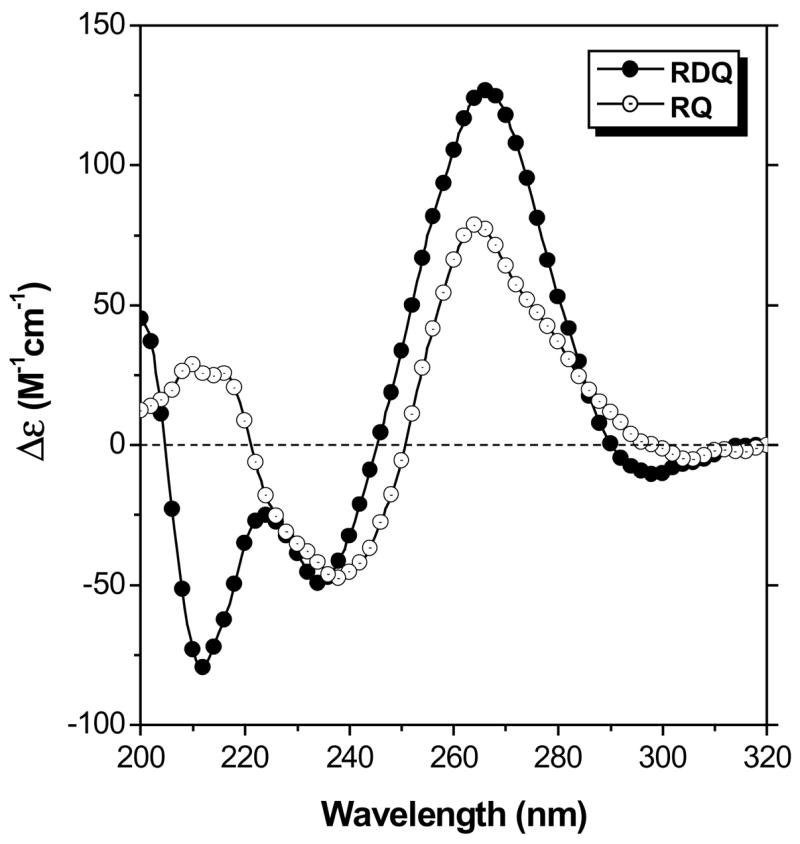
Meanwhile, the UV melting curves shown in Figure 2 illustrate how deletion of the stem simplifies thermal analysis of the RNA. The full length RNA exhibits two transitions when the absorbance is monitored at 295 nm. First, a hypochromic transition is observed with melting temperature (Tm) = ca. 45 °C. Further heating leads to a hyperchromic transition with Tm = ca. 80 °C. These transitions have been assigned to quadruplex melting at lower temperature to give a large hairpin structure followed by melting of the stem to produce the open, random coil RNA (29). This interpretation is supported by prior work showing that in DNA, melting of G-quadruplexes yields hypochromic transitions when monitored at 295 nm (40), while Watson-Crick duplexes exhibit hyperchromic transitions. Moreover, using LiCl instead of KCl in the buffer reduces the lower temperature transition in RDQ without affecting the higher temperature transition (29). This is consistent with the known preference of G-quadruplexes for K+ over Li+.
Figure 2.
UV melting curves recorded at λ = 295 nm for RDQ and RQ. [RNA] = 2.0 μM, [KCl] = 100 mM. Data were collected every 0.5 °C during a heating ramp of 1 °C/min.
Deletion of the stem results in a much simpler melting curve for RQ (Figure 2). A single hypochromic transition is observed at approximately the same temperature where the quadruplex domain in RDQ melts. The relatively clean transition allows thermodynamic parameters for quadruplex melting to be extracted from the curve (Table 1). Replacing KCl with LiCl in the buffer decreases the melting transition by ca. 20 °C (Supplemental Information, Figure S1), supporting the assignment of this transition to melting of a G-quadruplex.
Table 1.
Melting temperatures and thermodynamic parameters* for folding of RQ and hybridization with complementary (P7C) and homologous (P7H) PNAs.
| −ΔG298K (kcal/mol) | −ΔH (kcal/mol) | −TΔS298K (kcal/mol) | Tm (°C) | |
|---|---|---|---|---|
| RQ | 2.7 ± 0.5 | 39.5 ± 1.1 | 36.8 ± 1.0 | 47.2 ± 1.0 |
| RQ + P7C | 12.3 ± 0.4 | 43.0 ± 0.4 | 30.6 ± 2.0 | 58.7 ± 0.8 |
| RQ + 2P7H | 28.5 ± 0.4 | 95.7 ± 1.9 | 67.1 ± 2.0 | 70.1 ± 0.5 |
Determined by curve fitting (41).
While the topology of the G-quadruplex in RQ cannot be determined solely by the CD spectral shape and melting curve, the results presented above demonstrate that the RNA folds into a stable secondary structure based on stacked G-quartets and should serve as a suitable target for hybridization by complementary and homologous PNA probes.
Hybridization of Complementary PNA to RQ
The seven base PNA P7C is complementary to G13–G19 within RQ (numbering from the 5’-end and shown in blue in Chart 1). Hybridization of the PNA requires disruption of the quadruplex structure in order to form PNA-RNA base pairs with G13, G14, G18 and G19. Mixing P7C with RQ in equal concentrations yields the CD spectrum shown in Figure 3. The shape of the spectrum is similar to that of RQ alone, except at 210 nm. PNA-RNA duplexes exhibit CD maxima at 265 nm and minima at 240 nm (42), consistent with the spectrum shown in Figure 3.
Figure 3.
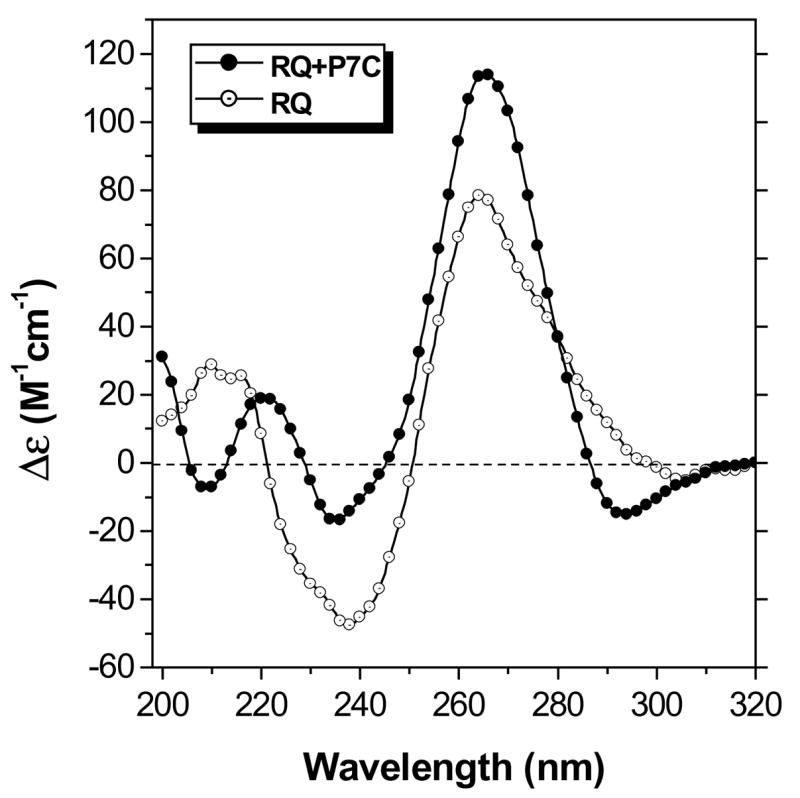
CD spectra recorded at 20 °C for RQ alone and with equimolar P7C. [RQ] = 2.0 μM, [KCl] = 100 mM.
Hybridization between P7C and RQ was also studied using a fluorescent labeled version of the PNA. Thiazole orange (TO) was linked to the PNA N-terminus to serve as a fluorescent reporter for hybridization (35). Upon hybridization with RQ, the TO dye should either stack on the end of the PNA-RNA hybrid or bind between unpaired bases on the RNA strand that extend beyond the PNA binding site. In either case, the conformational freedom of the dye will be reduced, leading to enhanced fluorescence (43,44). This behavior is not unique to PNA-linked TO. In fact, unsymmetrical cyanine dyes are well known for their enhanced fluorescence quantum yields when bound to DNA or RNA (45–47).
Figure 4 illustrates fluorescence emission spectra recorded for P7CTO alone in solution or in the presence of RQ. The fluorescence increases 27-fold in the presence of the RNA target, consistent with successful hybridization of the PNA. A continuous variations experiment verified that a 1:1 complex was formed (Figure S2) between the PNA and RNA. An experiment in which RQ was titrated into a solution of P7CTO was then performed with the fluorescence intensity recorded after each addition. The data were used to generate the binding isotherm shown in the inset to Figure 4, where fitting to a 1:1 binding model yielded an equilibrium constant (Kb) of 8.8 × 108 M−1cm−1. The same experiment performed using RDQ as the target yielded Kb = 8.1 × 108 M−1cm−1 (29), indicating that the stem of RDQ has almost no effect on the affinity of the PNA probe for the quadruplex target. These results illustrate that a short PNA oligomer can disrupt an RNA quadruplex secondary structure and form a PNA-RNA duplex at low nanomolar concentrations.
Figure 4.
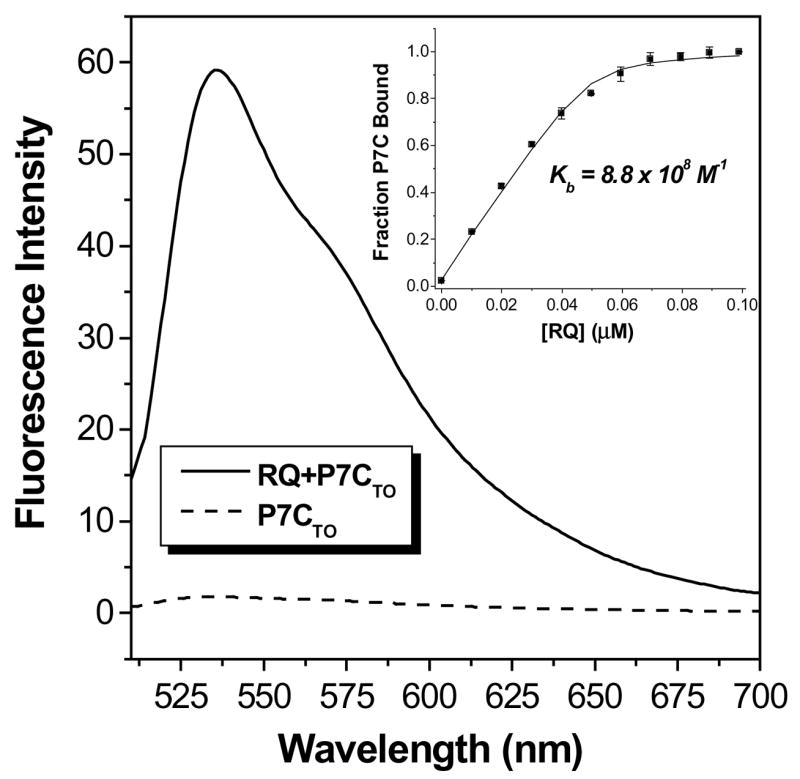
Fluorescence spectra recorded for P7CTO alone or with equimolar RQ. [PNA] = 2.0 μM, [KCl] = 100 mM. Samples were excited at 495 nm. Inset: Binding isotherm obtained from titrating RQ into P7CTO with fit to 1:1 binding model.
UV melting curves were recorded at two wavelengths (275 and 295 nm) to study PNA-RNA hybridization. Figure 5A shows that the hypochromic transition observed at 295 nm and assigned above to melting of the RQ quadruplex is lost in the presence of P7C, verifying that hybridization of the PNA disrupts the RNA secondary structure. Meanwhile, a hyperchromic transition is observed at 275 nm (Figure 5B) and this is assigned to denaturation of the PNA-RNA hybrid duplex. Since the melting temperature for the duplex is higher than that of the quadruplex, dissociation yields the unstructured RNA rather than re-folding of RQ into its quadruplex structure. Thermodynamic parameters for PNA-RNA hybridization were determined from the melting curve at 275 nm and are shown in Table 1. The equilibrium constant (Kb) calculated from the free energy change using the formula ΔG = −RTlnKb is 1.0 × 109 M−1, which matches well with that determined by fluorescence titration (Figure 4).
Figure 5.
UV melting curves recorded at 295 nm (A) and 275 nm (B) for RQ alone or with equimolar P7C. [RNA] = 2.0 μM, [KCl] = 100 mM. Data were collected every 0.5 °C during a heating ramp of 1 °C/min.
Hybridization of Homologous PNA to RQ
PNA P7H is homologous to the same region of RQ targeted by the complementary PNA P7C, i.e. G13–G19. With sufficient driving force, the PNA should be able to disrupt the RNA quadruplex and form a hybrid PNA-RNA quadruplex. In fact, prior work showed that P7H bound to RDQ in a 2:1 stoichiometry, meaning two PNAs associated with a single RNA (29). We speculated that both PNAs were forming quadruplexes, one with each “half” of the original RNA quadruplex. We sought to verify this using the simplified construct RQ.
Before performing hybridization experiments, we characterized the behavior of P7H in the absence of RNA. Guanine-rich PNAs have been shown to form homoquadruplex structures (48,49) and the extent to which this would interfere with P7H hybridization to RQ was unknown. The behavior of P7H in aqueous buffer was studied by CD and UV melting curves. A weak CD spectrum was recorded for the PNA but a relatively high concentration of the PNA (100 μM) was required (Figure S3A). At this concentration, a clear hypochromic transition was observed in the UV melting curve recorded at 295 nm (Figure S3B), indicating that P7H does form a homoquadruplex. However, at the lower concentrations used for the hybridization experiments (<5 μM) P7H exhibits neither a CD spectrum nor a UV melting transition. Therefore we consider the PNA to be monomeric under the conditions of the following experiments.
Figure 6 illustrates fluorescence spectra recorded for the TO-labeled analogue P7HTO alone and in the presence of RQ. The fluorescence enhancement is only 2.8-fold, compared with 27-fold for the complementary PNA targeted to the same RNA. The large difference in enhancement for the two probes is due to the background fluorescence of the unhybridized probe: the fluorescence is more than 10-fold higher for P7HTO than for P7CTO. The free probe fluorescence of TO-conjugated PNAs is highly dependent on the sequence and has been attributed to binding of the fluorophore within a folded or collapsed conformation of the PNA (50). The purine-rich P7HTO should be more hydrophobic than the pyrimidine-rich P7CTO, so it is not surprising that the G-rich PNA exhibits higher intrinsic fluorescence. A continuous variations experiment indicates formation of a 2:1 PNA-RNA hybrid (Figure 7), analogous to what was found previously with RDQ (29).
Figure 6.
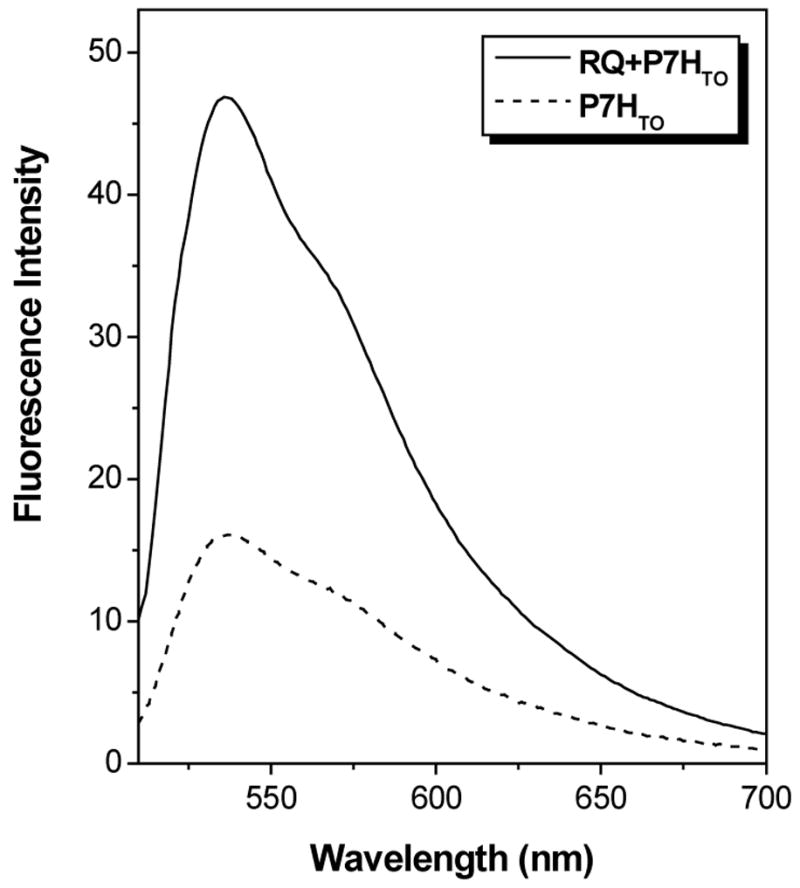
Fluorescence spectra recorded for P7HTO alone or with equimolar RQ. [PNA] = 2.0 μM, [KCl] = 100 mM. Samples were excited at 495 nm.
Figure 7.
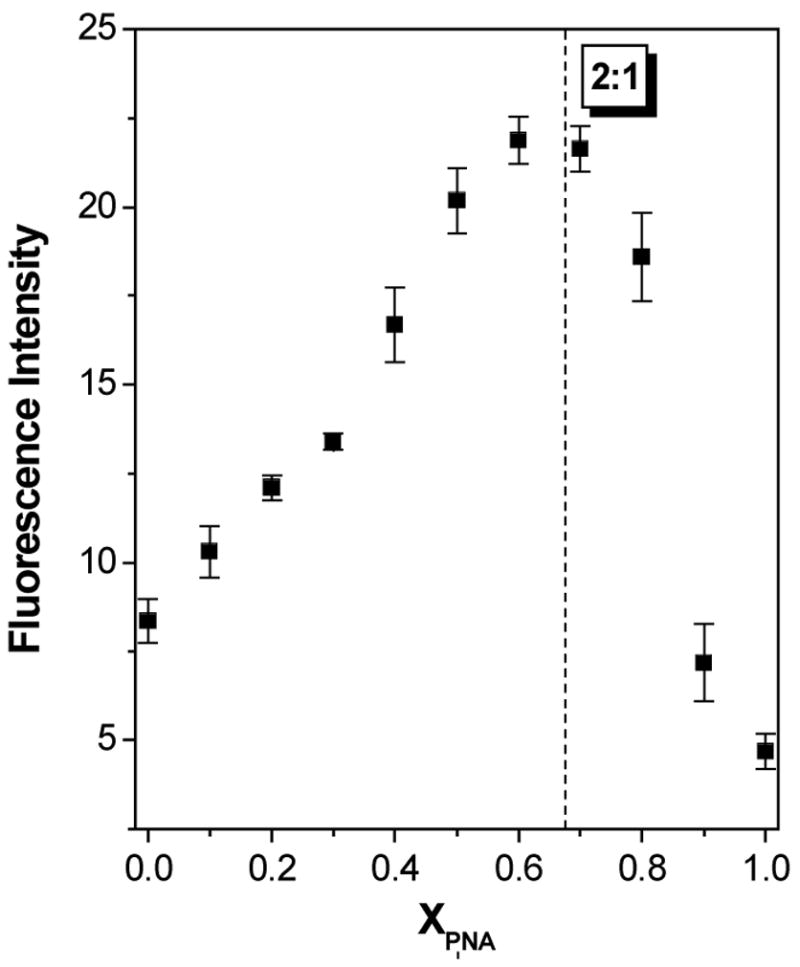
Continuous variations experiment to determine stoichiometry of P7HTO-RQ hybrid. Fluorescence intensity was recorded at 536 nm.
UV melting curves were used to further characterize the P7H-RQ hybrid. Figure 8 compares melting curves recorded at 295 nm for RQ alone and with one or two equivalents of the homologous PNA. In contrast to the complementary PNA, hybridization of RQ with P7H yields a complex that melts with a hypochromic transition (compare with Figure 5A). The transitions for the two PNA-containing samples occur at substantially higher temperatures than for RQ alone and the Tm is slightly higher when two equivalents of PNA are used. More striking is the variation in hypochromicity for the samples. The change in absorbance for RQ alone and with one equivalent of PNA are comparable but adding a second equivalent of PNA increases the hypochromicity by a factor of 2–3. Finally, substituting KCl with LiCl in the buffer decreases the Tm of the RQ-2P7H hybrid by ca. 40 °C (Figure 9), verifying that the complex consists of a hybrid quadruplex.
Figure 8.
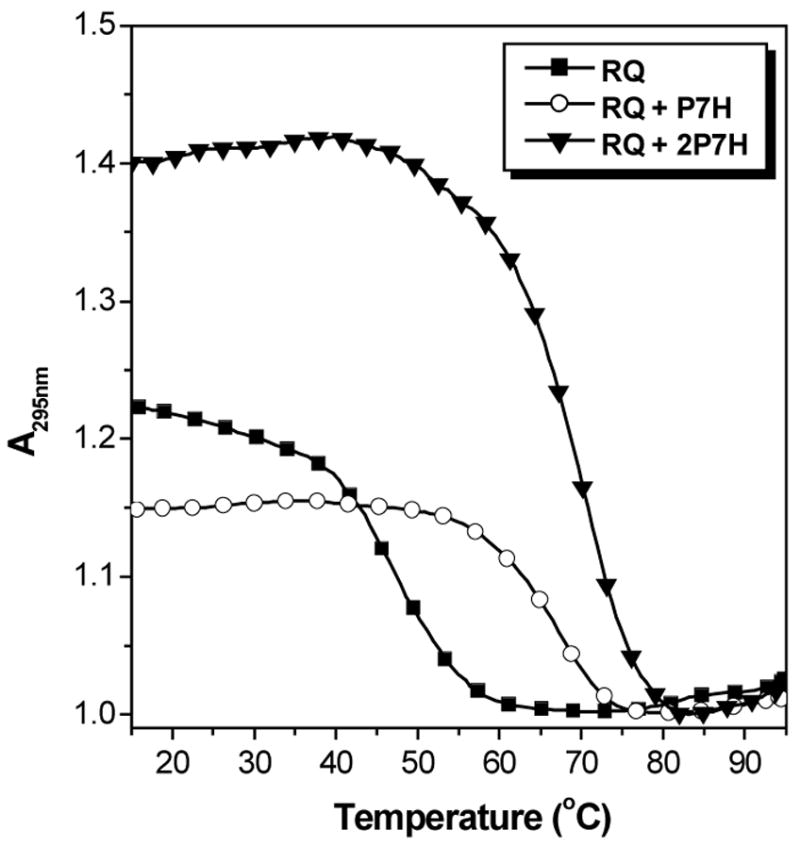
UV melting curves recorded at 295 nm for RQ alone or with one or two eq. P7H. [RNA] = 2.0 μM, [KCl] = 100 mM.
Figure 9.
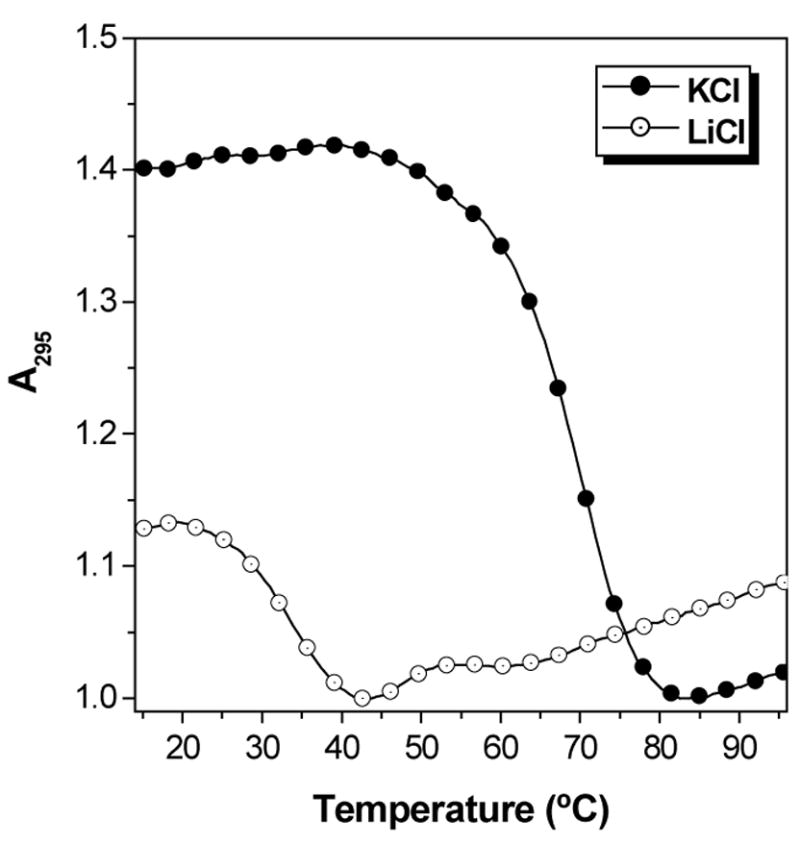
UV melting curves recorded at 295 nm for the RQ-2P7H hybrid. [RNA] = 2.0 μM, [KCl] = [LiCl] = 100 mM.
Thermodynamic parameters for hybridization of P7H with RQ were determined from the 2:1 UV melting curves shown above and are listed in Table 1. The hybrid quadruplex is much more stable than the RQ target (ΔG298K = −28.5 and −2.7 kcal/mol, respectively), explaining why the PNA easily disrupts the intramolecular quadruplex structure of RQ.
PNA Strand Orientation
While complementary hybridization of PNA with RNA is more favorable when the N-terminus of the PNA aligns with the 3’-rather than the 5’-terminus of the RNA (42), orientational preferences for hybridization of homologous PNA and RNA strands are unknown. Therefore, we synthesized the “reverse” homologous PNA in which the sequence is inverted to give P7Hr (Chart 1). This PNA hybridized with RQ to give CD spectra and UV melting curves that were nearly identical to those obtained with P7H-RQ (Figure 10), indicating that there is no orientational preference for hybrid quadruplex formation between RQ and short, homologous PNA oligomers.1
Figure 10.
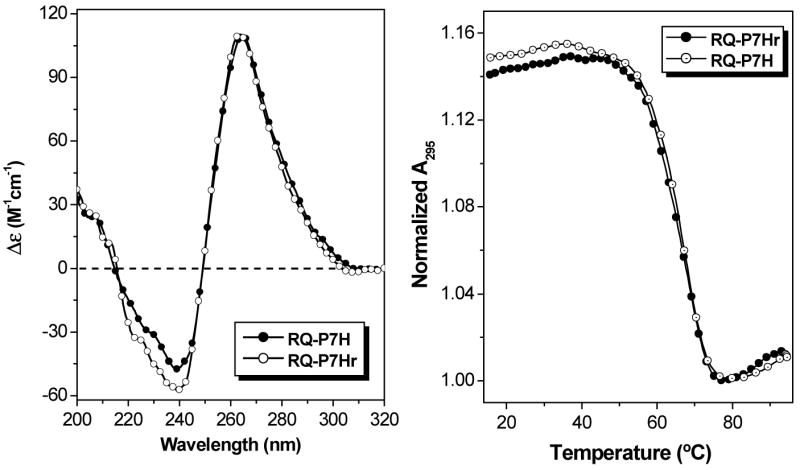
CD spectra (left) and UV melting curves recorded at 295 nm (right) for RQ-P7H and RQ-P7Hr hybrid quadruplexes. [RNA] = [PNA] = 2.0 μM, [KCl] = 100 mM.
DISCUSSION
The truncated RNA oligonucleotide RQ was used as a model for the full length RDQ we studied previously since deletion of the stem nucleotides was expected to simplify the UV melting curves. This was verified, as shown in Figure 2, where a single hypochromic transition corresponding to quadruplex melting is observed for RQ, compared with two transitions for RDQ. In addition to eliminating the high temperature stem-melting transition, deletion of the stem results in a ca. 3-fold larger hypochromicity than in the full length RNA, even though the melting temperatures are equivalent. The improvement in signal strength for RQ is most likely due to the deletion of 15 nucleotides from RDQ that absorb light at 295 nm but do not contribute to the hypochromicity.
One other aspect of the RNA targets is worthy of comment. The secondary structures shown in Chart 1 are based on previous literature reports for RDQ (12,14) and on the similarity of RQ to RDQ. The topologies of these structures are reminiscent of various DNA quadruplexes, including the thrombin-binding aptamer in which adjacent G-tracts have antiparallel orientations (19,20). The CD spectrum for that DNA exhibits a maximum at 295 nm (51). In contrast, the CD spectra for both RDQ and RQ exhibit maxima at 265 nm and minima at 240 nm. These features are similar to those exhibited by other G quadruplexes, such as that formed by a DNA model sequence based on a transcriptional regulatory region for the c-MYC oncogene (52). NMR spectroscopy has demonstrated that in the c-MYC quadruplex, adjacent G-tracts have parallel orientations, with the chain folding back twice on itself between successive G tracts via the loop nucleotides (i.e. the “double chain reversal”) (52,53). In another study, Oliver and Kneale reported that a DNA oligomer expected to form a parallel four-stranded quadruplex and its RNA homologue gave similar CD spectra, suggesting that the RNA formed a parallel structure as well (54). Thus, while CD spectra alone are not sufficient to determine strand orientations (55–57), these observations raise the possibility that RQ and RDQ adopt parallel rather than antiparallel folds.
Complementary versus Homologous Hybridization
Hybridization of the complementary PNA P7C to RQ yields a duplex structure with a CD spectrum similar to that reported previously for a 15 bp PNA-RNA hybrid (42). The melting temperature (Tm = 58.7 °C) and free energy change (ΔG298K = −12.3 kcal/mol) reflect not only the inherent high affinity of PNA for RNA, but also the stabilizing effect of overhanging nucleotides as well as the high G-C content of the duplex. In prior work, we hybridized a similar 7 base PNA (H-CCACACC-LysNH2) to the central 7 nucleotides of the quadruplex-forming DNA 5’-GGTTGGTGTGGTTGG-3’ and obtained similar hybrid stability: Tm = 55.2 °C and ΔG298K = −11.7 kcal/mol (27), although those measurements were performed at lower KCl concentrations (10 mM vs 100 mM). The top part of Figure 11 depicts the formation of the PNA-RNA hybrid duplex at the expense of the RNA quadruplex secondary structure.
Figure 11.
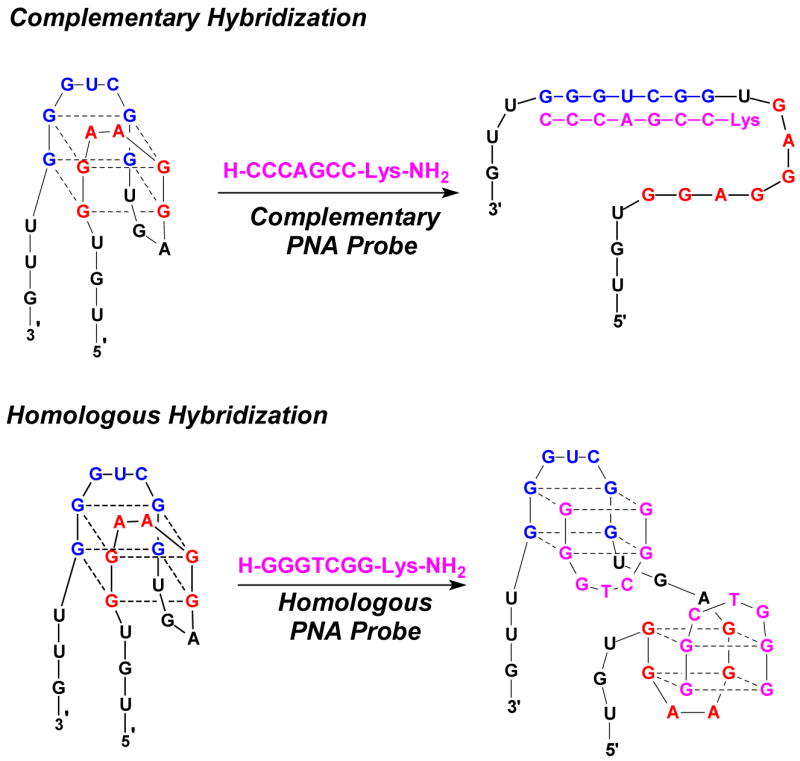
Comparison of complementary and homologous hybridization modes for PNA-RNA recognition. Note that there are several possible structures for the 2:1 hybrid quadruplex in addition to the one shown above.
Meanwhile, the homologous PNA P7H hybridizes to RQ to form a ternary PNA2-RNA quadruplex structure. This suggests that the PNA invades the RNA quadruplex and then binds to the two halves of the target. One possible structure is shown in the lower part of Figure 11, although the RNA quadruplex could also unfold by disrupting the hydrogen bonds between the two blue G-tracts and those between the two red G-tracts.
The thermodynamic parameters determined from the melting curve of the ternary complex are shown in Table 1 and indicate that formation of the 2:1 hybrid quadruplex is much more favorable enthalpically than formation of the 1:1 hybrid duplex when the complementary PNA P7C is used. This finding is due at least in part to the larger number of hydrogen bonds involved in formation of the 2:1 quadruplex (32 H-bonds) versus the duplex (20 H-bonds), although differences in base stacking, hydration and coordination of potassium ions by the G-tetrads likely contribute as well. Similarly, the additional loss of translational entropy incurred on formation of a ternary complex compared to a binary complex accounts for part of the more negative ΔS for the quadruplex. Conformational entropy is also an important consideration but more difficult to assess. While formation of the 1:1 duplex by P7C leaves unpaired overhangs of 3 and 11 nucleotides on the 3’ and 5’-termini, respectively, the 2:1 quadruplex formed by P7H leaves as many as 14 nucleotides unpaired on the RNA strand and 6 nucleotides on the two PNA strands (assuming no interaction between the loop bases). Regardless of how the contributions to the ΔH and ΔS are parsed, the hybrid quadruplex formed by P7H with RQ is very stable.
Hybrid quadruplex formation between homologous PNA and RNA strands is more complicated than hybrid duplex formation between complementary strands because there are at least four possible structures that could form (Figure 12). These vary not only in terms of strand alignment, i.e. whether the PNA N- or C-terminus aligns with the RNA 5’-terminus, but also with respect to the placements of the loops in the hybrid. We were able to assign strand orientations in the PNA2-DNA2 four-stranded quadruplex formed from the G4T4G4 sequence by a set of fluorescence resonance energy transfer experiments (30). Analogous experiments in the present system would be less informative due to the compact structure of both the target RQ and the hybrid quadruplex. However, we find that the CD spectrum of the hybrid PNA2-RNA quadruplex has a similar shape to the PNA2-DNA2 quadruplex, suggesting that the strand orientations are similar for the two hybrids. This implies alignment of the PNA N-terminus with the RNA 5’-terminus, eliminating the last two structures shown in Figure 12. Whether the RNA and PNA loops are on the same or opposite ends of the stacked G quartets cannot be determined at this time but will be the subject of future investigation. Note however that there is unlikely to be strong interaction between the loops, since inverting the orientation of the PNA (P7Hr vs P7H) has no discernable effect on the PNA2-RNA CD spectrum or melting temperature.
Figure 12.
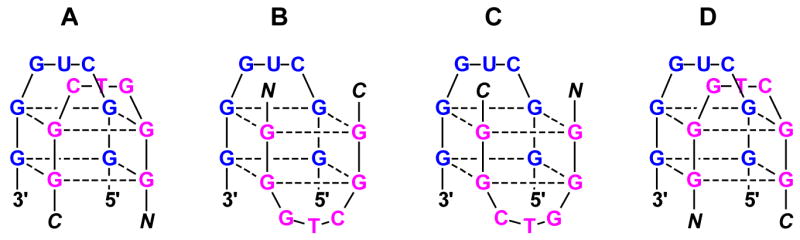
Four possible 1:1 hybrid quadruplexes formed by homologous RNA and PNA strands. In structures A and B, the PNA N-terminus aligns with the RNA 5’-terminus whereas the orientation is reversed for structures C and D.
Further Enhancements in Affinity
The results presented above illustrate that both complementary and homologous PNAs can hybridize with high affinity to G-quadruplex forming RNA targets. Further improvement in the affinity of complementary PNAs is straightforward and primarily involves lengthening the PNA so that additional base pairs can form. The homologous PNAs require a different approach if higher affinity is needed. One possibility is to covalently attach two homologous PNAs to reduce the translational entropic penalty of hybridization (58,59). Two considerations in designing such a bisPNA are (1) the positions at which the two PNA strands should be connected, and (2) preventing the bisPNA from folding into its own stable quadruplex. Structural analysis of a PNA2-RNA or PNA2-DNA quadruplex would be most helpful in addressing the first issue. The very low stability of the homoquadruplex formed by P7H suggests that even if a bimolecular quadruplex were to be formed by a bisPNA analogue, it should not significantly interfere with hybridization. Separately, quadruplex-binding small molecules could be attached to the PNA to “cap” the G-quartet stack, further stabilizing the hybrid quadruplex structure.
Finally, the ability of two homologous PNA strands to bind to RQ requires reconsideration of just what is meant by “homologous”. In the strictest definition, two homologous sequences are identical at every position. However, this is clearly unnecessary for P7H to bind to RQ since there is only one 7 nucleotide sequence within the RNA that is completely homologous to the PNA and yet two PNAs hybridize to the RNA. Rather, it seems that homology in the G-tracts is sufficient to permit hybrid quadruplex formation: RQ has four G2 tracts allowing two PNAs, each with two G2 tracts, to simultaneously hybridize. If there is no direct interaction between the loop nucleobases (i.e. those not involved in G-quartet formation), then the sequence selectivity of hybridization could suffer, since there are likely to be many potential binding sites consisting of two G2 tracts separated by a few nucleotides. Future experiments will focus on this issue by varying the number of nucleobases separating two G tracts within a given target and assessing hybrid quadruplex formation by G-rich PNAs.
Supplementary Material
Acknowledgments
We thank the National Institutes of Health (RO1 GM58547) and Carnegie Mellon University for financial support of this research, Dr. Babu Rao Renikuntla for synthesis of the thiazole orange derivative and Laurel Grotzinger for expert technical assistance. MALDI-TOF mass spectra were recorded in the Center for Molecular Analysis at Carnegie Mellon University, supported by NSF grants CHE-9808188 and DBI-9729351.
Footnotes
This work was funded by a grant from the National Institutes of Health (RO1 GM58547).
Interestingly, the complementary PNA-RNA duplex and homologous PNA-RNA quadruplex give similar CD spectra. However, the opposite melting curve profiles monitored at 295 nm readily distinguish between the two types of hybrids, as well as the dependence on cation identity.
References
- 1.Williamson JR. G-quartet structures in telomeric DNA. Annu Rev Biophys Biomol Struct. 1994;23:703–730. doi: 10.1146/annurev.bb.23.060194.003415. [DOI] [PubMed] [Google Scholar]
- 2.Simonsson T. G-quadruplex DNA structures - variations on a theme. Biol Chem. 2001;382:621–628. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- 3.Davis JT. G-quartets 40 years later: from 5'-GMP to molecular biology and supramolecular chemistry. Angew Chem Int Ed. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 4.Kumar N, Maiti S. Quadruplex to Watson-Crick duplex transition of the thrombin binding aptamer: a fluorescence resonance energy transfer study. Biochem Biophys Res Comm. 2004;319:759–767. doi: 10.1016/j.bbrc.2004.05.052. [DOI] [PubMed] [Google Scholar]
- 5.Williamson JR, Raghuraman MK, Cech TR. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 6.Shafer RH, Smirnov I. Biological aspects of DNA/RNA quadruplexes. Biopolymers. 2001;56:209–227. doi: 10.1002/1097-0282(2000/2001)56:3<209::AID-BIP10018>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 7.Catasti P, Chen X, Moyzis RK, Bradbury EM, Gupta G. Structure-function correlations of the insulin-linked polymorphic region. J Mol Biol. 1996;264:534–545. doi: 10.1006/jmbi.1996.0659. [DOI] [PubMed] [Google Scholar]
- 8.Simonsson T, Pecinka P, Kubista M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frantz JD, Gilbert W. A novel yeast gene product, G4p1, with a specific affinity for quadruplex nucleic acids. J Biol Chem. 1995;270:20692–20697. doi: 10.1074/jbc.270.35.20692. [DOI] [PubMed] [Google Scholar]
- 11.Schaeffer C, Bardoni B, Mandel JL, Ehresmann B, Ehresmann C, Moine H. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 13.Brown V, Jin P, Ceman S, Darnell JC, O’Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, Darnell RB, Warren ST. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in Fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 14.Ramos A, Hollingworth D, Pastore A. G-quartet-dependent recognition between the FMRP RGG box and RNA. RNA. 2003;9:1198–1207. doi: 10.1261/rna.5960503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sirand-Pugnet P, Durosay P, Brody E, Marie J. An intronic (A/U) GGG repeat enhances the splicing of an alternative intron of the chicken beta-tropomyosin pre-mRNA. Nucleic Acids Res. 1995;23:3501–3507. doi: 10.1093/nar/23.17.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomez D, Lemarteleur T, Lacroix L, Mailliet P, Mergny JL, Riou JF. Telomerase downregulation induced by the G-quadruplex ligand 12459 in A549 cells is mediated by hTERT RNA alternative splicing. Nucleic Acids Res. 2004;32:371–379. doi: 10.1093/nar/gkh181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonnal S, Schaeffer C, Créancier L, Clamens S, Moine H, Prats AC, Vagner S. A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons. J Biol Chem. 2003;278:39330–39336. doi: 10.1074/jbc.M305580200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 19.Macaya RF, Schultze P, Smith FW, Roe JA, Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc Natl Acad Sci USA. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang KY, McCurdy S, Shea RG, Swaminathan S, Bolton PH. A DNA aptamer which binds to and inhibits thrombin exhibits a new structural motif for DNA. Biochemistry. 1993;32:1899–1904. doi: 10.1021/bi00059a003. [DOI] [PubMed] [Google Scholar]
- 21.Wen J-D, Gray CW, Gray DM. SELEX selection of high-affinity oligonucleotides for bacteriophage Ff gene 5 protein. Biochemistry. 2001;40:9300–9310. doi: 10.1021/bi010109z. [DOI] [PubMed] [Google Scholar]
- 22.Andreola M-L, Pileur F, Calmels C, Ventura M, Tarrago-Litvak L, Toulmé J-J, Litvak S. DNA aptamers selected against the HIV-1 RNase H display in vitro antiviral activity. Biochemistry. 2001;40:10087–10094. doi: 10.1021/bi0108599. [DOI] [PubMed] [Google Scholar]
- 23.Mori T, Oguro A, Ohtsu T, Nakamura Y. RNA aptamers selected against the receptor activator of NF-kB acquire general affinity to proteins of the tumor necrosis factor receptor family. Nucleic Acids Res. 2004;32:6120–6128. doi: 10.1093/nar/gkh949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cuesta J, Read MA, Neidle S. The design of G-quadruplex ligands as telomerase inhibitors. Mini-Rev Med Chem. 2003;3:11–21. doi: 10.2174/1389557033405502. [DOI] [PubMed] [Google Scholar]
- 25.Seenisamy J, Bashyam S, Gokhale V, Vankayalapati H, Sun D, Siddiqui-Jain A, Streiner N, Shin-ya K, White E, Wilson WD, Hurley LH. Design and synthesis of an expanded porphyrin that has selectivity for the c-MYC G-quadruplex structure. J Am Chem Soc. 2005;127:2944–2959. doi: 10.1021/ja0444482. [DOI] [PubMed] [Google Scholar]
- 26.Rezler EM, Seenisamy J, Bashyam S, Kim MY, White E, Wilson WD, Hurley LH. Telomestatin and diseleno sapphyrin bind selectively to two different forms of the human telomeric G-quadruplex structure. J Am Chem Soc. 2005;127:9439–9447. doi: 10.1021/ja0505088. [DOI] [PubMed] [Google Scholar]
- 27.Datta B, Armitage BA. Hybridization of PNA to structured DNA targets: quadruplex invasion and the overhang effect. J Am Chem Soc. 2001;123:9612–9619. doi: 10.1021/ja016204c. [DOI] [PubMed] [Google Scholar]
- 28.Green JJ, Ying L, Klenerman D, Balasubramanian S. Kinetics of unfolding the human telomeric DNA quadruplex using a PNA trap. J Am Chem Soc. 2003;125:3763–3767. doi: 10.1021/ja029149w. [DOI] [PubMed] [Google Scholar]
- 29.Marin VL, Armitage BA. RNA guanine quadruplex invasion by complementary and homologous PNA probes. J Am Chem Soc. 2005;127:8032–8033. doi: 10.1021/ja051102y. [DOI] [PubMed] [Google Scholar]
- 30.Datta B, Schmitt C, Armitage BA. Formation of a PNA2-DNA2 hybrid quadruplex. J Am Chem Soc. 2003;125:4111–4118. doi: 10.1021/ja028323d. [DOI] [PubMed] [Google Scholar]
- 31.Dawson RMC, Elliott DC, Elliott WH, Jones KM. Data for Biochemical Research. Oxford University Press; New York: 1986. pp. 103–114. [Google Scholar]
- 32.Christensen L, Fitzpatrick R, Gildea B, Petersen KH, Hansen HF, Koch T, Egholm M, Buchardt O, Nielsen PE, Coull J, Berg RH. Solid-phase synthesis of peptide nucleic acids. J Peptide Sci. 1995;3:175–183. doi: 10.1002/psc.310010304. [DOI] [PubMed] [Google Scholar]
- 33.Koch T. In: Peptide Nucleic Acids: Protocols and Applications. Nielsen PE, editor. Horizon Bioscience; Norfolk: 2004. pp. 37–60. [Google Scholar]
- 34.Zhou X-f, Peng Z-h, Geise J, Peng B-x, Li Z-x, Yan M, Domisse R, Carieer R, Claeys M. Blue sensitizing dyes: synthesis, spectroscopy, and performance in photographic emulsions. J Imaging Sci Technol. 1995;39:244–252. [Google Scholar]
- 35.Svanvik N, Westman G, Wang D, Kubista M. Light-up probes: thiazole orange-conjugated peptide nucleic acid for detection of target nucleic acid in homogeneous solution. Anal Biochem. 2000;281:26–35. doi: 10.1006/abio.2000.4534. [DOI] [PubMed] [Google Scholar]
- 36.van Holde KE, Johnson CW, Ho PS. Principles of Physical Biochemistry. Prentice Hall; Upper Saddle River: 1998. p. 657. [Google Scholar]
- 37.Job P. Recherches sur la formation de complexes minéraux en solution, et sur leur stabilité. Ann Chim (Paris) 1928;9:113–203. [Google Scholar]
- 38.Dias N, Sénamaud-Beaufort C, le Forestier E, Auvin C, Hélène C, Saison-Behmoaras TE. RNA hairpin invasion and ribosome elongation arrest by mixed base PNA oligomer. J Mol Biol. 2002;320:489–501. doi: 10.1016/S0022-2836(02)00474-6. [DOI] [PubMed] [Google Scholar]
- 39.Gray DM, Tinoco IJ, Chamberlin MJ. The circular dichroism of synthetic ribonucleic acids and the influence of uracil on conformation. Biopolymers. 1972;11:1235–1258. doi: 10.1002/bip.1972.360110609. [DOI] [PubMed] [Google Scholar]
- 40.Mergny JL, Phan AT, Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 41.Marky LA, Breslauer KJ. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987;26:1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- 42.Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, Berg RH, Kim SK, Nordén B, Nielsen PE. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 43.Lee LG, Chen C, Liu LA. Thiazole orange: a new dye for reticulocyte analysis. Cytometry. 1986;7:508–517. doi: 10.1002/cyto.990070603. [DOI] [PubMed] [Google Scholar]
- 44.Nygren J, Svanvik N, Kubista M. The interactions between the fluorescent dye thiazole orange and DNA. Biopolymers. 1998;46:39–51. doi: 10.1002/(SICI)1097-0282(199807)46:1<39::AID-BIP4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 45.Rye HS, Yue S, Wemmer DE, Quesada MA, Haugland RP, Mathies RA, Glazer AN. Stable fluorescent complexes of double-stranded DNA with bis-intercalating asymmetric cyanine dyes: properties and applications. Nucleic Acids Res. 1992;20:2803–2812. doi: 10.1093/nar/20.11.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Netzel TL, Nafisi K, Zhao M, Lenhard JR, Johnson I. Base-content dependence of emission enhancements, quantum yields, and lifetimes for cyanine dyes bound to double-stranded DNA: photophysical properties of monomeric and bichromophoric DNA stains. J Phys Chem. 1995;99:17936–17947. [Google Scholar]
- 47.Ishiguro T, Saitoh J, Yawata H, Otsuka M, Inoue T, Sugiura Y. Fluorescence detection of specific sequence of nucleic acids by oxazole yellow-linked oligonucleotides. Homogeneous quantitative monitoring of in vivo transcription. Nucleic Acids Res. 1996;24:4992–4997. doi: 10.1093/nar/24.24.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krishnan-Ghosh Y, Stephens E, Balasubramanian S. A PNA4 quadruplex. J Am Chem Soc. 2004;126:5944–5945. doi: 10.1021/ja031508f. [DOI] [PubMed] [Google Scholar]
- 49.Datta B, Bier ME, Roy S, Armitage BA. Quadruplex formation by a guanine-rich PNA oligomer. J Am Chem Soc. 2005;127:4199–4207. doi: 10.1021/ja0446202. [DOI] [PubMed] [Google Scholar]
- 50.Svanvik N, Nygren J, Westman G, Kubista M. Free-probe fluorescence of light-up probes. J Am Chem Soc. 2001;123:803–809. doi: 10.1021/ja002294u. [DOI] [PubMed] [Google Scholar]
- 51.Smirnov I, Shafer RF. Effect of loop sequence and size on DNA aptamer stability. Biochemistry. 2000;39:1462–1468. doi: 10.1021/bi9919044. [DOI] [PubMed] [Google Scholar]
- 52.Seenisamy J, Rezler EM, Powell TJ, Tye D, Gokhale V, Joshi CS, Siddiqui-Jain A, Hurley LH. The dynamic character of the G-quadruplex element in the c-MYC promoter and modification by TMPyP4. J Am Chem Soc. 2004;126:8702–8709. doi: 10.1021/ja040022b. [DOI] [PubMed] [Google Scholar]
- 53.Phan AT, Modi YS, Patel DJ. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J Am Chem Soc. 2004:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oliver AW, Kneale GG. Structural characterization of DNA and RNA sequences recognized by the gene 5 protein of bacteriophage fd. Biochem J. 1999;339:525–531. [PMC free article] [PubMed] [Google Scholar]
- 55.Đapic V, Abdomerovic V, Marrington R, Peberdy J, Rodger A, Trent JO, Bates PJ. Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res. 2003;31:2097–2107. doi: 10.1093/nar/gkg316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Risitano A, Fox KR. Influence of loop size on the stability of intramolecular DNA quadruplexes. Nucleic Acids Res. 2004;32:2598–2606. doi: 10.1093/nar/gkh598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hazel P, Huppert J, Balasubramanian S, Neidle S. Loop-length-dependent folding of G-quadruplexes. J Am Chem Soc. 2004;126:16405–16415. doi: 10.1021/ja045154j. [DOI] [PubMed] [Google Scholar]
- 58.Griffith MC, Risen LM, Greig MJ, Lesnik EA, Sprankle KG, Griffey RH, Kiely JS, Freier SM. Single and bis peptide nucleic acids as triplexing agents: binding and stoichiometry. J Am Chem Soc. 1995;117:831–832. [Google Scholar]
- 59.Lesnik EA, Risen LM, Driver DA, Griffith MC, Sprankle K, Freier SM. Evaluation of pyrimidine PNA binding to ssDNA targets from nonequilibrium melting experiments. Nucleic Acids Res. 1997;25:568–574. doi: 10.1093/nar/25.3.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.