Figure 1.
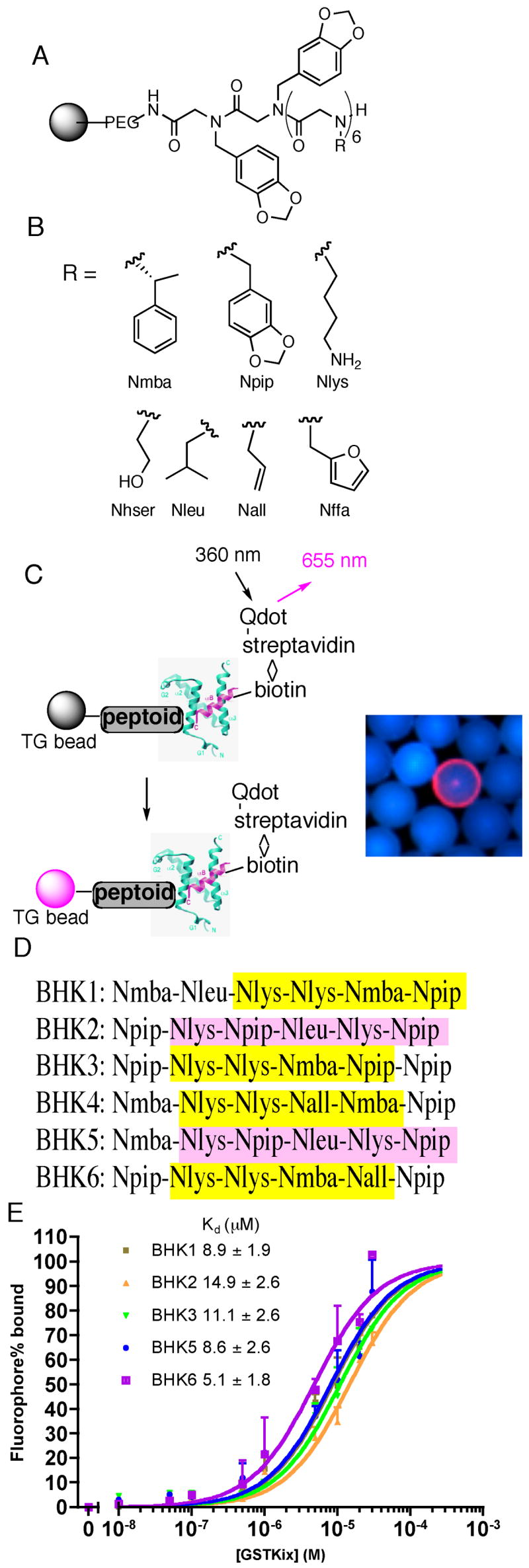
Isolation and characterization of KIX-binding peptoids BHK1-6. (A) General structure of the hexameric peptoid library. (B) The structure and code of the seven residues in the library. The codes are either the short abbreviations of the amines used in the synthesis (piperonylamine, (R)-(+)-α-methylbenzylamine, allylamine, furfurylamine) or derived from the side chain similarity with natural amino acids (lysine, leucine, homoserine). (C) Left: A schematic presentation of screening protocol utilizing red-emitting streptavidin-modified quantum dots. Right: A typical fluorescence micrograph of a bead scored as a hit (red halo) in the midst of large number of beads scored as negatives (blue). (D) The sequences of the six hits as determined by on-bead Edman degradation analysis (see Figure S1 for chemical structures). Similarities in the sequences are highlighted (see text for a discussion). (E) In vitro binding of carboxyfluorescein-substituted BHKs (see Figure S3 for structures) to GST-KIX monitored by fluorescence polarization. The observed polarization values (P) were converted to percentage of fluorophore binding [(P−Pmin)/(Pmax−Pmin) × 100%] using fluorophore only as Pmin, whereas Pmax was derived by fitting the raw P data to a one-site binding model in Prism4.0 (GraphPad Software, San Diego, CA). The apparent KDs, presented as mean ± SEM, were derived from the best fit of the data to the sigmoidal dose-response model in Prism4.0. Each data point, shown as mean + SD, represents an average of three independent measurements.
