Abstract
Eukaryotic cells under stress repress translation and localize these messenger RNAs (mRNAs) to cytoplasmic RNA granules. We show that specific stress stimuli induce the assembly of RNA granules in an organelle with bacterial ancestry, the chloroplast of Chlamydomonas reinhardtii. These chloroplast stress granules (cpSGs) form during oxidative stress and disassemble during recovery from stress. Like mammalian stress granules, cpSGs contain poly(A)-binding protein and the small, but not the large, ribosomal subunit. In addition, mRNAs are in continuous flux between polysomes and cpSGs during stress. Localization of cpSGs within the pyrenoid reveals that this chloroplast compartment functions in this stress response. The large subunit of ribulosebisphosphate carboxylase/oxygenase also assembles into cpSGs and is known to bind mRNAs during oxidative stress, raising the possibility that it plays a role in cpSG assembly. This discovery within such an organelle suggests that mRNA localization to granules during stress is a more general phenomenon than currently realized.
Introduction
Diverse eukaryotic cells under specific stress conditions repress the translation of many mRNAs and localize them to cytoplasmic RNA granules (Parker and Sheth, 2007; for review see Anderson and Kedersha, 2008). A particular class of these RNA granules in plants and mammals, the stress granules (SGs), form under stress caused by reactive oxygen species, UV light, high temperature, energy deprivation, and osmotic shock. They contain the small ribosomal subunit, translation initiation factors, poly(A)-binding protein (PABP), and other RNA-binding proteins. When SGs were first identified in plant cells, they were proposed to sequester translationally repressed mRNAs until the restoration of homeostasis (Nover et al., 1989). Mammalian SGs were shown subsequently to be a dynamic compartment in which mRNAs from disassembled polysomes are rapidly routed either back to polysomes, to processing bodies for degradation, or to RNPs for storage (Kedersha et al., 2000, 2005).
This study was initiated by our unexpected identification of RNA granules in the chloroplast of the green alga Chlamydomonas reinhardtii under a stress condition caused by exposure to high intensity light. We named them chloroplast SGs (cpSGs) because they are analogous to SGs (as reported here). In contrast to the nucleocytoplasmic genetic systems in which RNA granules have been found, chloroplast genomes are expressed by bacterial mechanisms. Chloroplast ribosomes have the sedimentation coefficient of bacterial ribosomes (70S), nearly a full complement of bacterial ribosomal proteins, and they use homologues of bacterial general translation factors (Beligni et al., 2004). Here we used fluorescence confocal microscopy and pharmacological approaches to characterize the composition of cpSGs, the stress conditions that induce their assembly, their location within the chloroplast, the mechanisms underlying their assembly, and the nature of the mRNAs that localize to them.
Results and discussion
Chloroplast mRNAs localize to cpSGs in cells under high light stress
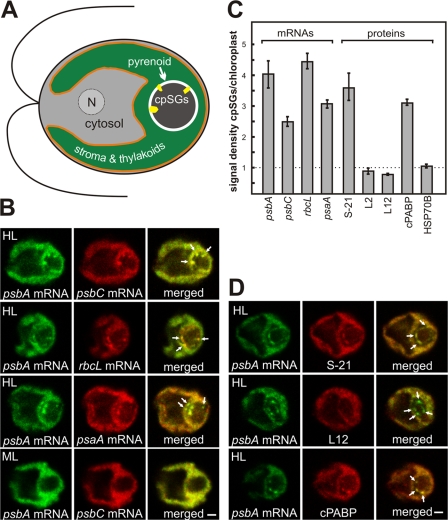
For each of the four chloroplast mRNAs examined by FISH and confocal microscopy, a fraction localized in cpSGs in most cells that were cultured under moderate intensity light and then exposed to high intensity light for 10 min (Fig. 1 B and see Fig. 2). This localization pattern was seen in a minority of cells maintained under moderate light. These chloroplast mRNAs encode subunits of photosystem II (PS II; psbA and psbC), photosystem I (psaA), and ribulosebisphosphate carboxylase/oxygenase (Rubisco; rbcL). This localization was quantified by determining the mean ratio of the fluorescence densities of each FISH signal in cpSGs versus in the chloroplast (n ≥ 20 cells for each mRNA). This analysis revealed that each of these mRNAs is concentrated severalfold in cpSGs (Fig. 1 C).
Figure 1.
cpSGs resemble cytoplasmic SGs in their composition of translation components. (A) An illustration based on the uppermost cell in B shows the chloroplast with cpSGs (yellow), the envelope (orange), the region with the aqueous stroma and thylakoid vesicles (green), and the pyrenoid (brown), surrounded by a starch sheath (white). Also shown are the cytosolic compartments (gray), the nucleus (N), and the flagella. (B) In cells under high light stress (HL), cpSGs had strong FISH signals from the mRNAs of psbA, psbC, rbcL, and psaA. cpSGs were not seen in most cells under the nonstress condition of moderate light (ML). (C) Bar heights indicate a mean signal density in cpSGs standardized to the signal density of the chloroplast for the FISH signals from the mRNAs and a protein of the small ribosomal subunit (S-21), two proteins of the large ribosomal subunit (L2 and L12), and cPABP. Error bars represent two SEM. A value substantially greater than one (dotted line) represents localization in cpSGs. (D) cpSGs revealed by the psbA FISH signal immunolabeled S-21, but only weakly for L12. cpSGs also showed strong immunofluorescence signal of cPABP. Arrows in B and D indicate cpSGs. The micrographs show 0.2-μm optical sections. Bars, 1 μm. For each experiment, n ≥ 20 cells.
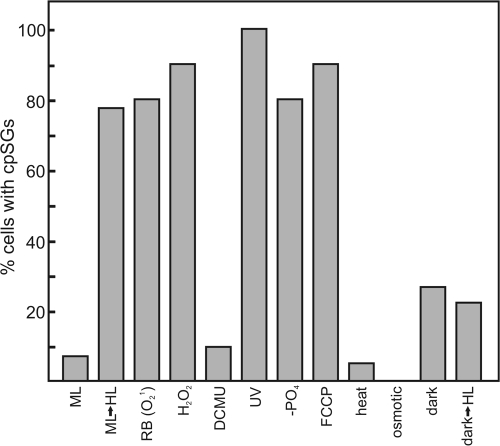
Figure 2.
cpSGs form under specific stress conditions. Bar heights indicate the percentages of cells with cpSGs under the following conditions: ML, constant moderate light; ML→HL, cells cultured under moderate light were exposed to high light; RB, oxidative stress was induced by rose bengal; H2O2, oxidative stress was induced by exposure to hydrogen peroxide; DCMU, a PS II inhibitor; UV, UV light exposure; −PO4, phosphate deprivation; FCCP, energy deprivation by this proton ionophore; heat, heat shock; osmotic, osmotic shock; dark, 2-h dark-adapted cells; dark→HL, 2-h dark-adapted exposed to HL. These data are representative of one experiment. For each condition, n ≥ 20 cells.
cpSGs and SGs are similar in their composition of translation components
To determine whether cpSGs are similar to SGs or other stress-induced RNA granules, we asked whether they contain the subunits of the chloroplast ribosome and the C. reinhardtii PABP homologue, cPABP (Yohn et al., 1998). Hallmarks of SGs are their enrichment for PABP and the small ribosomal subunit, but not the large ribosomal subunit. In cells from the high light stress condition, cpSGs were seen to have strong immunofluorescence from S-21, a protein of the small ribosomal subunit (Fig. 1 D). Quantification revealed a 3.5-fold higher signal density in cpSGs relative to the rest of the chloroplast (Fig. 1 C). Moreover, two proteins of the large ribosomal subunit, L12 and L2, had lower signal densities in cpSGs than in the chloroplast (Fig. 1, C and D; and see Fig. 3 C). Finally, the signal density of cPABP was threefold higher in cpSGs than in the chloroplast (Fig. 1, C and D). Although stable chloroplast mRNAs are not 3′ polyadenylated, genetic and biochemical evidence support a role of cPABP in chloroplast translation involving its binding to internal mRNA sequences (Yohn et al., 1998; Beligni et al., 2004). Thus, cpSGs resemble SGs in their composition of these translation components.
Figure 3.
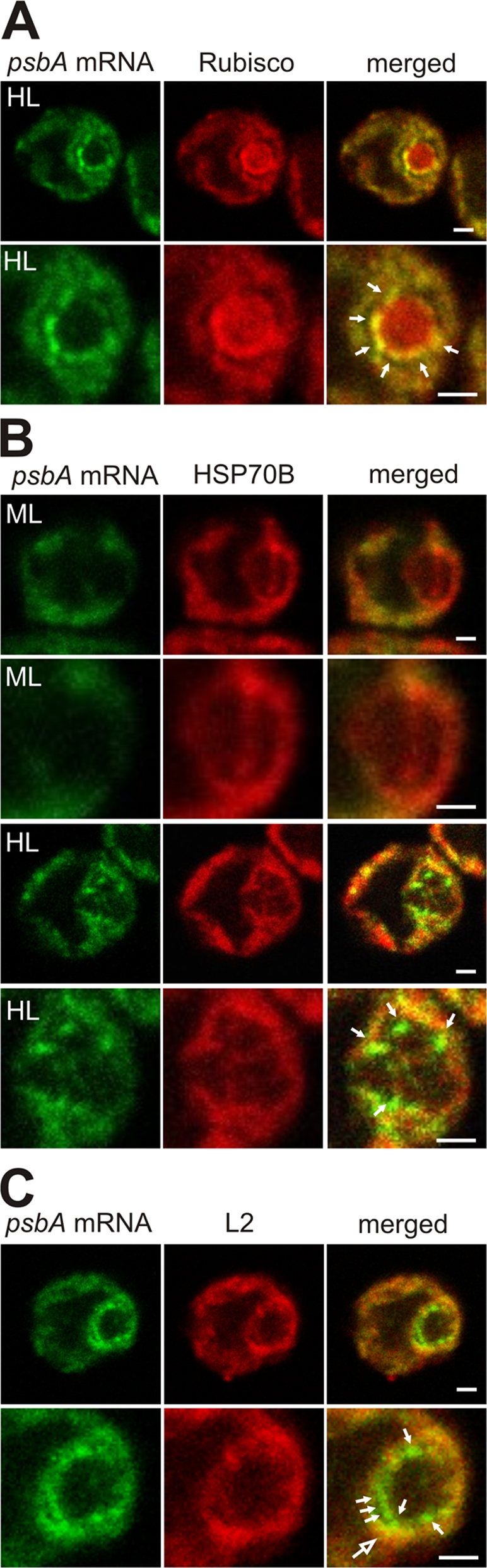
cpSGs are located at the internal perimeter of the pyrenoid. cpSGs, with the psbA mRNA (green), were located relative to proteins for the pyrenoid, stroma, and T zones. For each cell, a second image set shows an enlargement of the pyrenoid region. (A) In high light–stressed cells (HL), cpSGs were detected at the internal perimeter of the pyrenoid, revealed by immunolabeled Rubisco (100%, n = 20 cells). (B) In cells from both moderate and high light conditions, the chloroplast stroma marker protein HSP70B was observed in lines that appear to be pyrenoid tubules. The cpSGs in high light–stressed cells were detected at the peripheral termini of this HSP70B immunostaining pattern (n = 7 cells). (C) cpSGs are distinct from T zones (large arrow), which have colocalized psbA mRNA and ribosomal protein L2 (n = 20). Arrows indicate cpSGs and the larger arrow in C indicates a T zone. The micrographs show 0.2- μm optical sections. Bars, 1 μm.
Specific stress conditions induce mRNA localization to cpSGs
To analyze factors that induce mRNA localization to cpSGs, cells were exposed to different stresses and analyzed for the psbA mRNA distribution. Because high light exposure induces oxidative stress and damage to PS II (Murata et al., 2007), conditions specific to each of these stresses were tested. When oxidative stress was induced by treatment with hydrogen peroxide or rose bengal (a photosensitizer of singlet oxygen production; Fischer et al., 2004), higher percentages of cells with cpSGs were observed relative to nontreated cells (Fig. 2). In contrast, inhibition of PS II by 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) had no significant effect. Therefore, high light causes mRNA localization to cpSGs by inducing oxidative stress and not by damaging PS II.
Other results suggest that energy deprivation also induces mRNA localization to cpSGs. First, exposure to the proton ionophore carbonylcyanide-p-trifluoromethoxyphenylhydrazone (FCCP), which inhibits ATP synthesis in the chloroplasts and mitochondria, elevated the percentage of cells with cpSGs by more than 11-fold (Fig. 2). Second, a threefold increase occurred during a 2-h incubation in the dark, a condition that is unlikely to be associated with a stress condition apart from energy deprivation caused by the lack of photosynthesis. High light exposure of 2-h dark-adapted cells had little effect, possibly because they lacked polysomal psbA mRNAs for localization to cpSGs (see section mRNAs from disassembled polysomes localize to cpSGs; Fig. 2; Trebitsh et al., 2000). cpSGs were not described in an earlier paper, which examined dark-adapted cells under high light stress (Uniacke and Zerges, 2007). cpSG formation was also induced by exposure to UV light or phosphate deprivation. Heat shock had no effect and osmotic shock abolished cpSGs in the minority of cells that have them under moderate light (Fig. 2). Therefore, cpSGs form under specific stress conditions and the underlying inducing factors and signaling pathways remain to be determined.
cpSGs are located at the internal perimeter of the pyrenoid
To characterize the location of cpSGs, we used marker proteins for specific chloroplast compartments. cpSGs were detected only in the vicinity of the pyrenoid. This spherical compartment in the chloroplasts of most algae provides a high CO2/O2 ratio to favor the carboxylase activity of Rubisco in the Calvin cycle (Michael et al., 1991). In cells from the high light stress condition that were immunoprobed for the Rubisco holoenzyme to reveal the pyrenoid and FISH probed for the psbA mRNA, all cpSGs were located at the internal perimeter of the pyrenoid (Fig. 3 A). However, cpSGs were within patches of immunofluorescence from a marker protein for the chloroplast stroma HSP70B, where it was not enriched relative to its mean signal density in the chloroplast (Fig. 1 C and Fig. 3 B; Liu et al., 2005). Together, these results suggest that cpSGs are in pockets of stroma within the pyrenoid. Therefore, we considered whether cpSGs are within the opening of the membranous tubules that have been seen by EM to extend into the pyrenoid (Ohad et al., 1967). Consistent with this possibility, we observed cpSGs at the termini of what appeared to be pyrenoid tubules with HSP70B in rare images that longitudinally sectioned them (Fig. 3 B).
We proposed previously that discrete regions within the chloroplast, called T zones, house early steps of translation for de novo PS II assembly, based on the rapid colocalization therein of the psbA and psbC mRNAs and both subunits of the chloroplast ribosome specifically under conditions of de novo PS II assembly (Uniacke and Zerges, 2007). As T zones are near the pyrenoid, we compared their location with respect to cpSGs (see Materials and methods). As seen in merged images of the fluorescence signals from the psbA mRNA and L2 (Fig. 3 C), T zones (yellow) are distinct from cpSGs (green). Moreover, certain cpSGs adjoined a T zone, suggesting a functional relationship between these compartments.
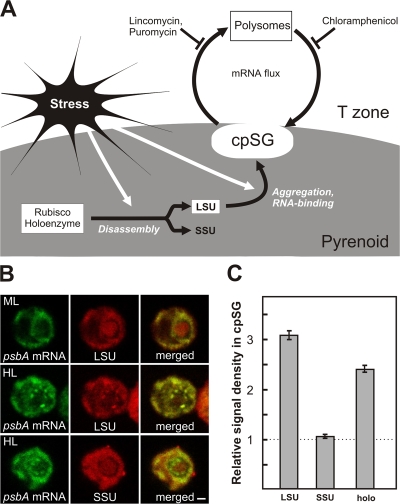
A model for cpSG assembly involving the large subunit of Rubisco (LSU)
A key difference between cpSGs and SGs are their factors for mRNA localization and assembly. SG assembly involves the self-aggregation of the RNA-binding proteins TIA-1 and TIAR (Anderson and Kedersha, 2008). Homologues were not identified in Basic Local Alignment Search Tool searches of the C. reinhardtii genome. However, a candidate cpSG assembly factor was suggested by our review of previous reports that C. reinhardtii cells under oxidative stress disassemble Rubisco into its constituent LSUs and small subunits (SSUs), whereupon the oxidized LSU nonspecifically binds chloroplast mRNAs and forms large particles (Yosef et al., 2004; Knopf and Shapira, 2005; Cohen et al., 2006). These authors proposed a role of LSU in the repression of the rbcL translation. In addition, immunogold EM revealed a particulate form of Rubisco at the internal periphery of the pyrenoid (Suss et al., 1995). Although this was proposed to function in the Calvin cycle, it may have been LSU in cpSGs.
To determine whether cpSG assembly involves the aggregation of LSU bound to chloroplast mRNAs (Fig. 4 A), we asked whether LSU, but not SSU, localizes to cpSGs. Indeed, when cells under high light stress were immunoprobed with antisera specific to each of these subunits, most of the LSU signal within the pyrenoid was in cpSGs, whereas the SSU signal remained dispersed (Fig. 4 B). Quantification of the mean signal densities in cpSGs, relative to the entire pyrenoid, confirmed that LSU localizes to cpSGs, whereas SSU does not (Fig. 4 C). Therefore, LSU could be an mRNA-binding factor in cpSG assembly, analogous to TIA-1 and TIAR in the assembly of SGs in mammalian cells (Anderson and Kedersha, 2008). Different assembly mechanisms probably would be involved because LSU lacks amino acid sequence similarity to the Q-rich prionlike domains in proteins that nucleate SG assembly (for review see Anderson and Kedersha, 2008).
Figure 4.
A model for cpSG assembly. (A) cpSG assembly is proposed to involve the stress-induced disassembly of the Rubisco holoenzyme in the pyrenoid, the activation of the RNA-binding activity of LSU, followed by its concurrent aggregation and binding to mRNAs that are released from polysomes (Knopf and Shapira, 2005; Cohen et al., 2006). Flux of mRNAs and small chloroplast ribosomal subunits between cpSGs and polysomes during stress was revealed by effects of the translation inhibitors. Lincomycin inhibits the initiation of protein synthesis, but allows ribosomes to complete translation, thereby liberating mRNA from polysomes. Puromycin liberates mRNAs from polysomes by inducing premature termination. Chloramphenicol prevents polysome disassembly by stalling translating ribosomes on mRNAs. (B) Cells from the nonstress condition of moderate light (ML) had immunolabeled LSU uniformly distributed within the pyrenoid. After the induction of stress by high light (HL), most LSU of the pyrenoid localized in cpSGs, whereas SSU remained dispersed. (C) To quantify the localization of Rubisco LSU, SSU, and holoenzyme to cpSGs, the mean density of each signal therein was standardized to its density throughout the pyrenoid. A value substantially greater than one (dotted line) represents localization in cpSGs. Error bars represent two SEM. The micrographs show 0.2-μm optical sections. Bar, 1 μm. For each experiment, n ≥ 20 cells.
mRNAs from disassembled polysomes localize to cpSGs
We addressed the nature of the mRNAs that localize to cpSGs. Cytoplasmic SGs contain mRNAs that are liberated by the disassembly of their polysomes resulting from translational repression, but not mRNAs that are highly translated during stress (Kedersha and Anderson, 2002; Stohr et al., 2006). There is evidence for translational repression of the rbcL and psbC mRNAs during high light stress (Cohen et al., 2006; Uniacke and Zerges, 2007). Although it is well known that high light stress activates psbA translation for the repair of photodamaged PS II complexes (Adir et al., 1990), a distinct psbA mRNA pool in T zones may become translationally repressed (Uniacke and Zerges, 2007). Although these results are consistent with the localization of mRNAs from disassembled polysomes to cpSGs, the following experiments were required to answer this question.
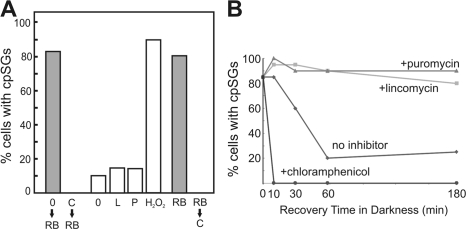
In mammalian cells, evidence that SGs receive mRNAs from disassembled polysomes was provided by studies involving pharmacological inhibition of translation. SG assembly was prevented by inhibitors of translation elongation, which trap mRNAs on polysomes. Similarly, treatment of C. reinhardtii cells with chloramphenicol, an inhibitor of translation elongation in the chloroplast, prevented cpSG assembly when oxidative stress was induced subsequently by rose bengal (Fig. 5 A). Moreover, under these conditions, LSU was not detected in cpSGs, suggesting that the mRNAs released by polysome disassembly are required either to trigger LSU aggregation or to serve as a structural component of cpSGs (unpublished data; n = 20).
Figure 5.
Evidence for psbA mRNA flux between cpSGs and polysomes during stress. (A) The percentage of cells with cpSGs is graphed for moderate light cells incubated without or with chloramphenicol and then exposed to rose bengal to induce oxidative stress (shaded bars; 0→RB and C→RB, respectively). Under nonstress conditions, mRNA release from polysomes only marginally induced cpSG formation as revealed by the percentages of cells with cpSGs that were exposed to no inhibitor (0), lincomycin (L), or puromycin (P), with treatment with hydrogen peroxide (H2O2) as a positive control. The cpSGs that formed in response to oxidative stress induced by exposure to rose bengal were abolished by a chloramphenicol treatment (shaded bars; RB and RB→C, respectively). (B) During recovery from rose bengal–induced oxidative stress in the dark, 60 min was required for the percentage of cells with cpSGs to drop to that of dark-adapted cells in the absence of inhibitor (diamonds). cpSG disassembly was accelerated by chloramphenicol (circles) and prevented by both lincomycin (squares) and puromycin (triangles). These data are representative of one experiment, n ≥ 20 cells for each.
In mammalian cells under nonstress conditions, SGs formed when mRNAs were liberated from polysomes by translation initiation inhibitors (Kedersha et al., 2000; Mazroui et al., 2006). However, if cpSG formation involves stress-induced Rubisco disassembly and aggregation of LSU, mRNAs released from polysomes in the absence of stress should not localize to cpSGs. Alternatively, if RNA-binding active LSU or any other protein that localizes mRNAs to cpSGs is always available, mRNAs released from polysomes under nonstress conditions should localize to cpSGs. Therefore, polysome disassembly in the chloroplast was induced by lincomycin or puromycin, which inhibit translation initiation and induce premature termination, respectively. In the absence of inhibitor under these conditions (see Materials and methods), 10% of cells had cpSGs (Fig. 5 A). In the presence of either inhibitor, 15% of the cells had cpSGs, a marginal increase compared with the effects of most stress conditions (Fig. 2). Results of a positive control experiment revealed that most of these cells could form cpSGs upon exposure to hydrogen peroxide. Therefore, in addition to mRNAs from polysome disassembly, cpSG assembly requires a stress condition, e.g., to induce Rubisco disassembly and activate LSU for RNA binding (Fig. 4 A).
mRNA flux occurs between cpSGs and polysomes
In mammalian cells, mRNA flux between SGs and polysomes was demonstrated, in part, by the rapid disappearance of SGs when mRNAs were trapped in the ribosome-bound pool by inhibitors of translation elongation during sustained oxidative stress (Kedersha et al., 2000). Similar mRNA dynamics appear to occur in the C. reinhardtii chloroplast. First, during sustained oxidative stress induced by rose bengal, the 80% of cells that had formed cpSGs lost them during a 10-min treatment with chloramphenicol, an inhibitor of elongation by chloroplast ribosomes (Fig. 5 A). Second, chloramphenicol dramatically accelerated the disappearance of cpSGs during recovery from oxidative stress (Fig. 5 B). Third, lincomycin and puromycin prevented cpSG disappearance within 180 min of recovery from stress, suggesting that mRNAs must be engaged by translating ribosomes to leave cpSGs. These inhibitors also initially enhanced the percentage of cells with cpSGs, presumably because they released mRNAs from polysomes in the minority of cells that had not done so in response to oxidative stress (Fig. 5 B). Thus, like mammalian SGs, cpSGs do not sequester mRNAs during stress. Rather, mRNAs are in continuous flux between polysomes and cpSGs (Fig. 4 A). Such mRNA dynamics could be facilitated by the proximity of cpSGs to T zones, where the psbA and psbC mRNAs are translated (Fig. 3 C; Uniacke and Zerges, 2007).
In conclusion, our discovery of cpSGs reveals a novel chloroplast stress response and, to our knowledge, the first example of a stress-induced RNA granule in a bacterial lineage or organellar genetic system. The location of cpSGs suggests that the pyrenoid provides an environment depleted of reactive oxygen species, which are produced during stress throughout the rest of the chloroplast and known to damage RNA (Nishiyama et al., 2006). Our results raise the possibility of a novel function of Rubisco LSU as an mRNA-localizing and assembly factor of cpSGs. The RNA-binding activity of LSU and its activation by oxidizing conditions are conserved in higher plants, suggesting that we have identified a general chloroplast stress response (Cohen et al., 2006). It remains to be determined whether cpSGs and SGs evolved from a common ancestral RNA granule or by convergent evolution from distinct origins in response to general requirements of mRNAs released from polysomes during stress. In either case, a comprehensive understanding of cpSGs should elucidate these requirements and the molecular mechanisms that fulfill them in chloroplasts.
Materials and methods
C. reinhardtii culture conditions
Strain CC-503 was cultured in high salt minimal medium (Sueoka, 1960) to a cell density of 2–3 × 106 cells/ml at 24°C under a light intensity of 150 μE m−1 s−2 and with orbital shaking. High light (2,000 μE m−1 s−2) was generated by a slide projector (Kodak), to which small (2–30-ml) cultures were exposed at a distance of 10 cm for 10 min with manual shaking. Dark adaption was a 2-h incubation of liquid cultures wrapped with two layers of aluminum foil on an orbital shaker. Lincomycin and chloramphenicol were added to a final concentration of 200 μg/ml. Puromycin was added to a final concentration of 1 mM. These inhibitors were shown to be active because each abolished protein synthesis in the chloroplast after 10 min as revealed by the results of in vivo radioisotope-pulse labeling experiments. Cells were treated with rose bengal or hydrogen peroxide at concentrations of 0.5 μM and 2 mM, respectively, for 15 min. DCMU was added to a final concentration of 10 μM for 15 min. FCCP was added to a final concentration of 10 μM for 30 min. UV irradiation (20 mJ) was performed in a Stratalinker (Promega) and cells were fixed 30 min later. The conditions of heat shock, osmotic shock, and phosphate deprivation were described previously (von Gromoff et al., 1989; Hoffmann and Beck, 2005; Moseley et al., 2006). For the condition shown in Fig. 3 C, the colocalization of the psbA mRNA and the chloroplast ribosomal protein L2 to T zones was induced by exposing 2-h dark-adapted cells to moderate light for 5 min as described previously (Uniacke and Zerges, 2007). These cells were then treated with lincomycin under moderate light, when it does induce cpSGs by potentiating light-induced stress (Hideg et al., 2007). For the experiments shown in Fig. 5 A, the effects of these inhibitors were assessed in the dark to avoid the induction of light stress. These cells were first cultured under moderate light (so they would have polysomes from which chloroplast mRNAs could be released) and then shifted to the dark immediately before the addition of lincomycin or puromycin.
FISH and immunofluorescence staining
The procedures and probes used for FISH and immunofluorescence were described previously (Colon-Ramos et al., 2003; Uniacke and Zerges, 2007). High specificity of the psbA, rbcL, and psaA FISH signals were demonstrated previously (Uniacke and Zerges, 2007). Indirect immunofluorescence was carried out using the following primary antisera: anti-SSU, anti-LSU (N. Brisson, University of Montreal, Montreal, Canada), anti-L2, anti-S-21, anti-Rubisco (E. Harris, Duke University, Durham, NC), anti-cPABP (S. Mayfield, The Scripps Research Institute, La Jolla, CA), and anti-HSP70B (M. Schroda, University of Freiburg, Freiburg, Germany). The in situ immunofluorescence staining patterns of the ribosomal proteins, PABP, and HSP70B were highly specific because each of the antibodies detected a protein of the expected molecular weight on immunoblots of total cellular protein. Secondary antisera alone did not generate a signal.
Confocal microscopy
Confocal images were obtained as 0.2-μm optical sections with a confocal laser-scanning microscope (TCS SP2; Leica) and image acquisition software (version 2.61; Leica). Argon and green helium neon lasers were used to produce the 488- and 543-nm stimulation of TRITC or the fluorophores on the FISH probes Alexa 488 and 555 (Invitrogen). The cells were observed with immersion oil using an HCX PL APO objective lens of 100× and NA of 1.4. Images were acquired in 512 × 512 format with digital zoom set at 4.8× with a pinhole size of 0.84 airy. Images were acquired after adjusting the maximal signal in each section to just below saturation. Fluorescence density was measured with the Measure tool of ImageJ.
Acknowledgments
We thank M. Champagne, P. Lasko, R. Mazroui, and J. Scrivens for critical review of the manuscript, B. Forster and M. Shapira for helpful comments, and N. Brisson, E. Harris, S. Mayfield, and M. Schroda for generous gifts of antisera.
This study used the confocal microscope of the Centre for Structural and Functional Genomics (Concordia University) and was funded by the Natural Sciences and Engineering Council of Canada grant 217566-03.
Abbreviations used in this paper: cpSG, chloroplast SG; DCMU, 3-(3,4-dichlorophenyl)-1,1-dimethylurea; FCCP, carbonylcyanide-p-trifluoromethoxyphenylhydrazone; LSU, large subunit of Rubisco; PABP, poly(A)-binding protein; PS II, Photosystem II; Rubisco, ribulosebisphosphate carboxylase/oxygenase; SG, stress granule; SSU, small subunit of Rubisco.
References
- Adir, N., S. Shochat, and I. Ohad. 1990. Light-dependent D1 protein synthesis and translocation is regulated by reaction center II. Reaction center II serves as an acceptor for the D1 precursor. J. Biol. Chem. 265:12563–12568. [PubMed] [Google Scholar]
- Anderson, P., and N. Kedersha. 2008. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33:141–150. [DOI] [PubMed] [Google Scholar]
- Beligni, M.V., K. Yamaguchi, and S.P. Mayfield. 2004. The translational apparatus of Chlamydomonas reinhardtii chloroplast. Photosynth. Res. 82:315–325. [DOI] [PubMed] [Google Scholar]
- Cohen, I., Y. Sapir, and M. Shapira. 2006. A conserved mechanism controls translation of Rubisco large subunit in different photosynthetic organisms. Plant Physiol. 141:1089–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colon-Ramos, D.A., J.L. Salisbury, M.A. Sanders, S.M. Shenoy, R.H. Singer, and M.A. Garcia-Blanco. 2003. Asymmetric distribution of nuclear pore complexes and the cytoplasmic localization of beta2-tubulin mRNA in Chlamydomonas reinhardtii. Dev. Cell. 4:941–952. [DOI] [PubMed] [Google Scholar]
- Fischer, B.B., A. Krieger-Liszkay, and R.L. Eggen. 2004. Photosensitizers neutral red (type I) and rose bengal (type II) cause light-dependent toxicity in Chlamydomonas reinhardtii and induce the Gpxh gene via increased singlet oxygen formation. Environ. Sci. Technol. 38:6307–6313. [DOI] [PubMed] [Google Scholar]
- Hideg, E., P.B. Kos, and I. Vass. 2007. Photosystem II damage induced by chemically generated singlet oxygen in tobacco leaves. Physiol. Plant. 131:33–40. [DOI] [PubMed] [Google Scholar]
- Hoffmann, X.K., and C.F. Beck. 2005. Mating-induced shedding of cell walls, removal of walls from vegetative cells, and osmotic stress induce presumed cell wall genes in Chlamydomonas. Plant Physiol. 139:999–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha, N., and P. Anderson. 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30:963–969. [DOI] [PubMed] [Google Scholar]
- Kedersha, N., M.R. Cho, W. Li, P.W. Yacono, S. Chen, N. Gilks, D.E. Golan, and P. Anderson. 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151:1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha, N., G. Stoecklin, M. Ayodele, P. Yacono, J. Lykke-Andersen, M.J. Fritzler, D. Scheuner, R.J. Kaufman, D.E. Golan, and P. Anderson. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopf, J.A., and M. Shapira. 2005. Degradation of Rubisco SSU during oxidative stress triggers aggregation of Rubisco particles in Chlamydomonas reinhardtii. Planta. 222:787–793. [DOI] [PubMed] [Google Scholar]
- Liu, C., F. Willmund, J.P. Whitelegge, S. Hawat, B. Knapp, M. Lodha, and M. Schroda. 2005. J-domain protein CDJ2 and HSP70B are a plastidic chaperone pair that interacts with vesicle-inducing protein in plastids 1. Mol. Biol. Cell. 16:1165–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui, R., R. Sukarieh, M.E. Bordeleau, R.J. Kaufman, P. Northcote, J. Tanaka, I. Gallouzi, and J. Pelletier. 2006. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Mol. Biol. Cell. 17:4212–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael, R., L. McKay, and S.P. Gibbs. 1991. Composition and function of pyrenoids: cytochemical and immunocytochemical approaches. Can. J. Bot. 69:1040–1052. [Google Scholar]
- Moseley, J.L., C.W. Chang, and A.R. Grossman. 2006. Genome-based approaches to understanding phosphorus deprivation responses and PSR1 control in Chlamydomonas reinhardtii. Eukaryot. Cell. 5:26–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata, N., S. Takahashi, Y. Nishiyama, and S.I. Allakhverdiev. 2007. Photoinhibition of photosystem II under environmental stress. Biochim. Biophys. Acta. 1767:414–421. [DOI] [PubMed] [Google Scholar]
- Nishiyama, Y., S.I. Allakhverdiev, and N. Murata. 2006. A new paradigm for the action of reactive oxygen species in the photoinhibition of photosystem II. Biochim. Biophys. Acta. 1757:742–749. [DOI] [PubMed] [Google Scholar]
- Nover, L., K.D. Scharf, and D. Neumann. 1989. Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol. Cell. Biol. 9:1298–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad, I., P. Siekevitz, and G.E. Palade. 1967. Biogenesis of chloroplast membranes. II. Plastid differentiation during greening of a dark-grown algal mutant (Chlamydomonas reinhardi). J. Cell Biol. 35:553–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, R., and U. Sheth. 2007. P bodies and the control of mRNA translation and degradation. Mol. Cell. 25:635–646. [DOI] [PubMed] [Google Scholar]
- Stohr, N., M. Lederer, C. Reinke, S. Meyer, M. Hatzfeld, R.H. Singer, and S. Huttelmaier. 2006. ZBP1 regulates mRNA stability during cellular stress. J. Cell Biol. 175:527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueoka, N. 1960. Mitotic replication of deoxyribonucleic acid in Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. USA. 46:83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suss, K.H., I. Prokhorenko, and K. Adler. 1995. In situ association of Calvin cycle enzymes, ribulose-1,5-bisphosphate carboxylase/oxygenase activase, ferredoxin-NADP+ reductase, and nitrite reductase with thylakoid and pyrenoid membranes of Chlamydomonas reinhardtii chloroplasts as revealed by immunoelectron microscopy. Plant Physiol. 107:1387–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebitsh, T., A. Levitan, A. Sofer, and A. Danon. 2000. Translation of chloroplast psbA mRNA is modulated in the light by counteracting oxidizing and reducing activities. Mol. Cell. Biol. 20:1116–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uniacke, J., and W. Zerges. 2007. Photosystem II assembly and repair are differentially localized in Chlamydomonas. Plant Cell. 19:3640–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gromoff, E.D., U. Treier, and C.F. Beck. 1989. Three light-inducible heat shock genes of Chlamydomonas reinhardtii. Mol. Cell. Biol. 9:3911–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohn, C.B., A. Cohen, A. Danon, and S.P. Mayfield. 1998. A poly(A) binding protein functions in the chloroplast as a message-specific translation factor. Proc. Natl. Acad. Sci. USA. 95:2238–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef, I., V. Irihimovitch, J.A. Knopf, I. Cohen, I. Orr-Dahan, E. Nahum, C. Keasar, and M. Shapira. 2004. RNA binding activity of the ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit from Chlamydomonas reinhardtii. J. Biol. Chem. 279:10148–10156. [DOI] [PubMed] [Google Scholar]