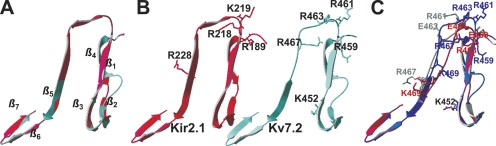
Figure 11.
Homology modeling of the helix A-B linker of Kv7.2 based on a domain of Kir2.1 as a template predicts a similar structure. Residues 428–484 of Kv7.2 were modeled using the program Swiss-Model using residues 186–245 of the solved crystal structure of Kir2.1 (Pegan et al., 2005) as a template (see Materials and methods). (A) Shown superimposed are the structure of the Kir2.1 domain (red) with that predicted for Kv7.2 (blue). The seven predicted β-sheets in the Kv7.2 domain are indicated. (B) The two structures are shown individually, with the basic residues identified in this study as being critical for PIP2 interactions with Kv7.2, and the corresponding residues by sequence alignment in Kir2.1 displayed in stick rendering. (C) Shown superimposed are the predicted structures of the wt Kv7.2 linker (blue), the Kv7.2 KRR-EEE triple mutant (red), and the Kv7.2 R463E mutant (gray), with the critical residues displayed in stick rendering. Note the alteration in predicted structure at the top of the linker induced by the mutations.