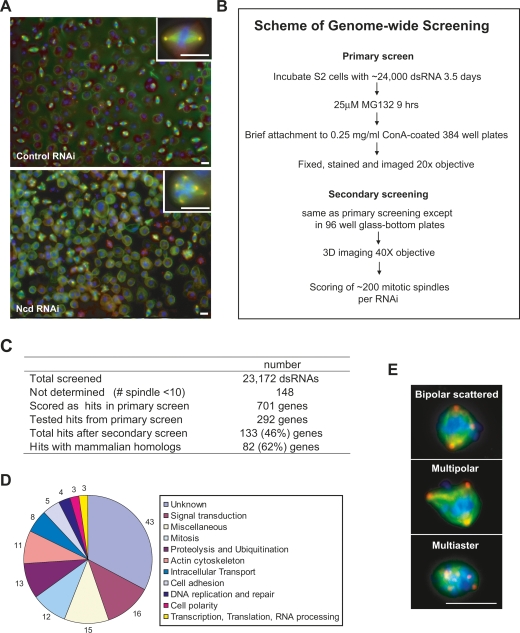
Figure 1.
Genome-wide screen for genes required for clustering supernumerary centrosomes in S2 cells. (A) Sample images from screen. Normal bipolar spindles in control EGFP RNAi (top panel) and multipolar spindles in Ncd RNAi (bottom panel) are shown. Cells were stained for MTs (α-tubulin, green), centrosomes (γ-tubulin, red), and DNA (blue). (Insets) Images at high magnification. (B) Scheme of genome-wide screen in S2 cells. Procedure for the primary and secondary screens. Addition of MG132, a proteasome inhibitor, at the end of the RNAi treatment was used to increase mitotic index. Immediate transfer of cells to Con-A was then used to facilitate cell attachment. (C) Table summarizing the screen results. (D) Gene Ontology (GO) annotations for the 133 genes from the screen. (E) Images showing classes of centrosome clustering defects in S2 cells. Cells were stained for MTs (α-tubulin, green), centrosomes (γ-tubulin, red), and DNA (blue). Bar, 10 μm.