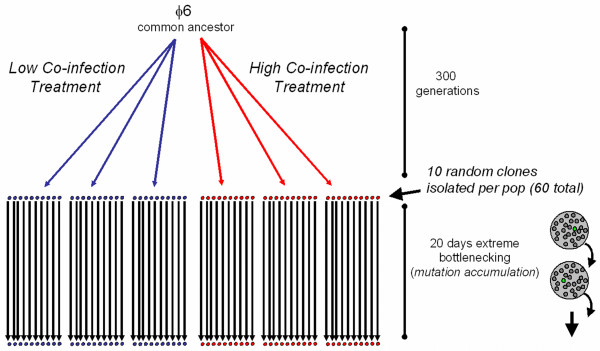
Figure 2.
Design for an evolution experiment where a wild type bacteriophage φ6 ancestor was used to found 3 lineages in a low level of co-infection treatment, and 3 lineages in a high co-infection treatment. After 60 days (300 generations), 10 clones were isolated at random from each population, and used to found lineages that were subjected to a mutation accumulation experiment. Cartoon at lower right depicts plaque-to-plaque transfers, where propagating the lineage through extreme bottlenecks of a single individual (green plaque) causes drift to overwhelm selection. The mutation-accumulation study was used to reveal whether prior ecological history (low versus high co-infection) affected the evolution of robustness: virus ability to maintain a constant phenotype (fitness) in the face of random mutational change.