Abstract
The molecular genetics of inherited phaeochromocytoma have received considerable attention, but the somatic genetic and epigenetic events that characterise tumourigenesis in sporadic phaeochromocytomas are less well defined. Previously, we found considerable overlap between patterns of promoter region tumour suppressor gene (TSG) hypermethylation in two neural crest tumours, neuroblastoma and phaeochromocytoma. In order to identify candidate biomarkers and epigenetically inactivated TSGs in phaeochromocytoma and neuroblastoma, we characterised changes in gene expression in three neuroblastoma cell lines after treatment with the demethylating agent 5-azacytidine. Promoter region methylation status was then determined for 28 genes that demonstrated increased expression after demethylation. Three genes HSP47, homeobox A9 (HOXA9) and opioid binding protein (OPCML) were methylated in >10% of phaeochromocytomas (52, 17 and 12% respectively). Two of the genes, epithelial membrane protein 3 (EMP3) and HSP47, demonstrated significantly more frequent methylation in neuroblastoma than phaeochromocytoma. These findings extend epigenotype of phaeochromocytoma and identify candidate genes implicated in sporadic phaeochromocytoma tumourigenesis.
Introduction
Neuroblastoma and phaeochromocytoma are the commonest neural crest-derived tumours in children and adults respectively. Most phaeochromocytomas are benign catecholamine-producing tumours arising within the adrenal medulla, but may be extra-adrenal (when they may be designated as paragangliomas) and/or approximately malignant (Chrisoulidou et al. 2007, Disick & Palese 2007). Up to one-third of phaeochromocytomas occur in genetically susceptible individuals (Astuti et al. 2001a,b, Neumann et al. 2002, Gimenez-Roqueplo 2006). Inherited predisposition to phaeochromocytoma may be associated with mutations in NF1, RET, SDHB, SDHC, SDHD or VHL genes, but somatic mutations in VHL, RET, NF1, SDHB and SDHD are rare in sporadic phaeochromocytomas (Eng et al. 1995, Hofstra et al. 1996, Astuti et al. 2001b,c, 2003). Promoter region hypermethylation and transcriptional silencing is a frequent cause of tumour suppressor gene (TSG) inactivation in many human cancers. Previously, in order to identify candidate epigenetically inactivated TSGs in phaeochromocytoma, we analysed promoter methylation status in a series of candidate genes known to undergo epigenetic silencing and found similar patterns of promoter hypermethylation in neuroblastoma and phaeochromocytoma. Thus, TSP1, CASP8, HIC1, DcR1, DcR2 and DR4 and RASSF1 were methylated frequently in both tumour types (Astuti et al. 2001c, Margetts et al. 2005). In contrast to phaeochromocytoma, familial neuroblastoma is rare. However, the genetic and epigenetic events associated with tumourigenesis in sporadic neuroblastoma are better defined than in phaeochromocytoma and molecular investigations of phaeochromocytoma tumourigenesis have been hampered by the absence of a human phaeochromocytoma cell line.
Based on our previous studies, we hypothesised that the identification of novel genes exhibiting promoter methylation in neuroblastoma would also provide plausible candidate genes for phaeochromocytoma tumourigenesis. In order to expand the number of candidate ‘epigenetically silenced TSGs’, we investigated a functional epigenomic approach (Alaminos et al. 2005) in neuroblastoma cell lines in order to identify candidate epigenetically inactivated TSGs in neuroblastoma and phaeochromocytoma.
Patients and methods
Patients and samples
DNA from a total of 52 tumour samples were analysed (19 NB's, 19 VHL-associated phaeochromocytomas and 14 sporadic phaeochromocytomas). Informed consent and approval from the appropriate Institutional Review Boards were obtained for all samples. DNA was extracted by standard methods. Normal human adult adrenal cDNA and genomic DNA were obtained from AMS Biotechnology (Europe) Ltd (Abingdon, Oxon, UK).
Cell lines
The ten neuroblastoma cell lines used were SK-N-AS, SK-N-F1, SK-N-DZ, SK-N-MC, SK-N-BE, SK-N-SH, NMB and LAN-5 (American Type Culture Collection, Manassas, VA, USA), KELLY and CHP212.
Sodium bisulphate modification
Sodium bisulphite modification was carried out using an adapted method (Herman et al. 1996). Genomic DNA (0.5–1.0 μg) was denatured at 37 °C for 10 min in 0.3 M NaOH. Unmethylated cytosines were sulphonated by incubation in 3.12 M sodium bisulphite and 1 M hydroquinone (pH 5) at (95 °C (30 s) 50 °C (15 min)) for 20 cycles. The resulting sulphonated DNA was purified using the Wizard DNA clean-up system (Promega), according to the manufacturer's instructions, except that DNA was eluted with distilled water (50 μl) at room temperature. Following elution, DNA was desulphonated in 0.3 M NaOH for 5 min at room temperature, then the DNA was precipitated with NaOAc (5 μl of 3 M) and ethanol (125 μl of 100%) overnight at −20 °C and resuspended in 50 μl distilled water.
Direct bisulphate DNA sequencing and methylation-specific PCR (MSP)
We determined CpG island methylation status by direct sequencing of bisulphite-modified genomic DNA or MSP.
For direct bisulphite sequencing, PCR products were excised from agarose gels and extracted using the QIAquick Gel Extraction kit (Qiagen), according to the manufacturer's instructions. Products were confirmed by direct sequencing from the forward or reverse PCR primer using ABI PRISM BigDye Terminator v3.1 Cycle sequencing kit (Genpak Ltd, Brighton, Sussex, UK) according to the manufacturer's instructions and run using ABI PRISM 3700 automatic sequencer (See Supplementary Table 1 which can be viewed online at http://erc.endocrinology-journals.org/supplemental/ for primers and PCR conditions).
MSP was performed essentially using previously published primers and conditions (Conway et al. 2000, Sellar et al. 2003, Yang et al. 2004, Alaminos et al. 2005, Breault et al. 2005, Lind et al. 2006). Expected PCR products are as follows: for HSP47(a), 134 (methylated)/135 (unmethylated) bp; HSP47(b), 109 (methylated)/110 (unmethylated) bp; EMP3, 252 (methylated)/255 (unmethylated) bp; opioid binding protein (OPCML), 135 (methylated)/135 (unmethylated) bp; CTNNG, 95 (methylated)/85 (unmethylated) bp; SYK, 242 (methylated)/140 (unmethylated); TMS1, 212 (methylated)/209 (unmethylated) bp; Homeobox A9 (HOXA9), 127 (methylated)/139 (unmethylated) bp (see Supplementary Table 1 which can be viewed online at http://erc.endocrinology-journals.org/supplemental/).
Reactions were hot-started at 95 °C for 15 min, by using 0.25 μl (5 units/μl) of HotstarTaq DNA polymerase (Qiagen). PCR products were visualised on 2% agarose gels stained with ethidium bromide. Genomic DNA methylated in vitro using SssI methylase (New England Biolabs, Ipswich, MA, USA) was used as a positive control for MSP and direct bisulphite sequencing.
Treatment of cell lines with 5-aza-2′-deoxycytidine
5-aza-2′-deoxycytidine (5-aza-dC, Sigma) was freshly prepared in ddH2O at 2 mg/ml and filter sterilised. Cells (1×106) were plated in 75 cm2 flask in RPMI 1640 medium supplemented with 10% FCS and left to settle for 24 h (day 0). Cells were treated with 2 μM of 5-aza-dC at days 1 and 4 and harvested at day 5. The culture medium was changed before each treatment and 24 h after treatment. Total RNA was extracted using the RNeasy kit (Qiagen) according to the manufacturer's guidelines.
U133A Plus Affymetrix oligonucleotide array analysis of neuroblastoma cell lines and gene expression analysis
Total RNA was isolated from the 5-aza-dC-treated and untreated neuroblastoma cell lines (SK-N-F1, SK-N-BE and SK-N-DZ) using the RNeasy Mini kit (Qiagen) and subsequently cleaned using the RNeasy mini columns (Qiagen) according to manufacturer's instructions. The quality and integrity of the RNA was verified by checking 28S and 18S rRNA after ethidium bromide staining of total RNA samples on 1% agarose gel electrophoresis. cDNA was performed using the SuperScript Double Stranded cDNA Synthesis kit (Invitrogen). cRNA was synthesised by in vitro transcription with biotinylated UTP and CTP. Labelled nucleic acid target quality was assessed by test 2 arrays and hybridised (45 °C for 16 h) to Affymetrix Human U133A plus oligonucleotide arrays. After automated washing and staining, absolute values of expression were calculated and normalised from the scanned array by using Affymetrix Microarray Suite (Version 5.0; Santa Clara, CA, USA). The Affymetrix RNA microrray was performed by Light Laboratories (Dr E Smith), University of Leeds using standard procedures.
Gene expression analysis for individual genes was performed by reverse transcription-PCR (RT-PCR). One microgram of RNA was reverse transcribed using Reverse Transcription Systems and oligo dT primers (Promega) according to the manufacturer's protocols. One microlitre of the cDNA obtained was then used as template for PCR amplification. Primer sequences and conditions are detailed in Supplementary Table 2, which can be viewed online at http://erc.endocrinology-journals.org/supplemental/. As a control, the GAPDH primers used were: 5′-AAGGTGAAGGTCGGAGTCAACG-3′ and 5′-CAGCCTTCTCCATGGTGGTGAA-3′, resulting in a PCR product of 319 bp. PCR products were visualised on 2% agarose gels stained with ethidium bromide.
Statistical analysis
Fisher's exact test was used as appropriate. P values of <0.05 were taken as statistically significant.
Results
Identification and evaluation of differentially expressed genes after demethylation of three neuroblastoma cell lines
Following treatment with the demethylating agent 5-aza-dC (5 μmol/l) for 5 days to reactivate the epigenetically silenced/downregulated genes, gene expression patterns pre- and post-treatment were compared using Gene Expression Microarrays (Affymetrix HG-U133A) in three neuroblastoma cell lines (SK-N-F1, SK-N-BE and SK-N-DZ). Candidate epigenetically silenced genes were prioritised for possible further investigation if a) they were known to be expressed in normal adrenal tissue (http://genome.ucsc.edu/cgi-bin/hgGateway), b) they were not located on the X chromosome or known to be imprinted, c) there was a CpG island proximal to the transcriptional start site that contained a predicted promoter sequence and d) treatment with 5-aza-dC was associated with a >2-fold upregulation, in at least two cell lines (or one cell line for genes previously implicated in human tumourigenesis).
Evaluation of candidate gene selection criteria for genes known to be epigenetically inactivated in neural crest tumours
Previously, we identified promoter region methylation in neuroblastoma and phaeochromocytoma for DcR1, DcR2, DR4, CASP8 and TSP1. To evaluate our candidate gene selection criteria, we investigated the correlation between promoter methylation status and fold changes in expression for these five genes in the three cell lines. Generally, there was a good correlation e.g. DcR1, which is methylated in SK-N-F1 and SK-N-DZ, but not in SK-N-BE was associated with microarray expression changes of 2.77-, 2.47- and 0.47-fold respectively. If the proposed selection criteria had been applied to these five genes then four (DcR1, DcR2, DR4 and CASP8) would have been selected for further analysis. We then applied our criteria and selected 27 candidate genes for further investigation (Table 1). Prior to performing 5′ CpG island promoter methylation, we evaluated the validity of the gene expression microarray analysis results for 10 genes (K19, Stannin (SNN), RASD1, actin-related protein 2/3 complex, sub unit 4 (ARPC4), protein tyrosine kinase 2 beta (PTK2B), transcription factor AP-4 (TFAP4), Zinc finger protein 36, C3H type-like 2 (ZFP36L2), ARGHDIA, suppressor of lin-12-like (SEL1) and latent-transforming growth factor β-binding protein 3 (LTBP3)) by RT-PCR analysis before and after 5-aza-dC treatment in each of the three neuroblastoma cell lines. In each case, expression was consistent with the results of microarray analysis (data not shown).
Table 1.
Data from microarray analysis showing candidate genes with greatest fold changes in expression after treatment with 5-aza-dC. Genes in bold were already known to have a role in the pathogenesis of neuroblastomas
| Affy ID | Gene | Gene name | SK-N-F1 | SK-N-BE | SK-N-DZ |
|---|---|---|---|---|---|
| 201650_at | KRT19 | Keratin 19 | 1.95 | 3.73 | 63.71 |
| 218033_s_at | SNN | Stannin | 3.76 | 1.00 | 50.50 |
| 206215_at | OPCML | Opioid binding protein | 20.52 | 3.52 | 0.26 |
| 223467_at | RASD1 | Activator of G protein signalling (AGS1) | 19.64 | 3.03 | 0.65 |
| 217817_at | ARPC4 | Actin-related protein 2/3 complex, sub unit 4 | 11.99 | 1.02 | 3.98 |
| 203110_at | PTK2B | Protein tyrosine kinase 2 beta | 2.73 | 10.95 | 2.02 |
| 205688_at | TFAP4 | Transcription factor AP-4 | 2.45 | 7.45 | 4.96 |
| 201367_s_at | ZFP36L2 | Zinc finger protein 36, C3H type-like 2 | 2.98 | 2.96 | 8.71 |
| 201167_x_at | ARHGDIA | ρ GDP dissociation inhibitor (GDI) α | 5.51 | 1.50 | 3.88 |
| 231248_at | CST6 | Cystatin E/M | 2.73 | 2.5 | 5.62 |
| 202062_s_at | SEL1L | Suppressor of lin-12-like | 2.64 | 2.97 | 5.12 |
| 219922_s_at | LTBP3 | Latent-transforming growth factor β-binding protein 3 | 0.82 | 3.60 | 5.17 |
| 209427_at | SMTH | Smoothelin | 2.91 | 2.59 | 3.88 |
| 205346_at | ST3GAL2 | ST3 β-galactoside α-2,3-sialyltransferase 2 | 3.77 | 1.43 | 3.71 |
| 201015_s_at | JUP | Junction plakoglobin/CTNNG | 1.44 | 3.08 | 3.50 |
| 209878_s_at | RELA | v-rel reticuloendotheliosis viral oncogene homologue A | 1.73 | 3.32 | 2.74 |
| 209905_at | HOXA9 | Homeobox A9 | 0.94 | 2.71 | 4.13 |
| 204911_s_at | TRIM3 | Tripartite motif-containing 3 | 2.07 | 1.49 | 4.09 |
| 217250_s_at | CHD5 | Chromodomain helicase DNA -binding protein 5 | 0.06 | 3.36 | 3.9 |
| 208325_s_at | PRKA2R | A kinase (PRKA) anchor protein 13 | 0.85 | 3.94 | 2.30 |
| 202588_at | AK1 | Adenylate kinase 1 | 2.43 | 1.53 | 2.79 |
| 203729_at | EMP3 | Epithelial membrane protein 3 | 2.11 | 2.94 | 1.61 |
| 208997_s_at | UCP2 | Uncoupling protein 2 | 2.09 | 1.79 | 2.48 |
| 207740_s_at | NUP62 | Nucleoporin 62 kDa | 2.39 | 0.93 | 2.82 |
| 222650_s_at | SLC2A4 | SLC2A4 regulator | 1.14 | 2.38 | 2.44 |
| 207540_s_at | SYK | Spleen tyrosine kinase | 0.33 | 3.76 | 0.78 |
| 207714_s_at | SERPINH1 | Heat shock protein 47 | 2.85 | 1.58 | 1.59 |
| 221666_s_at | TMS1 | Target of methylation-induced silencing 1 | 1.27 | 1.41 | 20.08 |
| 201781_s_at | AIP | Aryl hydrocarbon receptor-interacting protein | 1.01 | 0.86 | 2.29 |
5′ CpG methylation status for candidate epigenetically inactivated genes
After applying the proposed selection criteria, 21 candidate ‘novel epigenetically silenced TSGs’ were selected according to the strict criteria (upregulated in two or more cell lines) and a further seven genes previously reported to be methylated in other tumour types and upregulated in at least one cell line were selected for further analysis.
The methylation status of the 5′ promoter region CpG island was determined by direct sequencing of bisulphite-converted DNA. Out of the 21 candidate novel phaeochromocytoma TSGs, 19 (excepting KRT19 and CST6) were completely unmethylated in the three neuroblastoma cell lines and in adult normal adrenal DNA. Sequencing of the Keratin 19 (KRT19), 5′ CpG island revealed heavy methylation in all three cell lines (22/29, 25/29 and 29/29 CpG dinucleotides examined were methylated in SK-N-F1, SK-N-BE and SK-N-DZ respectively). However, the normal adrenal DNA also showed a similar pattern of CpG methylation (24/29 CpG dinucleotides methylated). Direct bisulphite sequencing of CST6 (Cystatin M) 5′ promoter region revealed methylated CpGs in all three neuroblastoma cell lines (data not shown). The CpG island methylation was also detected in four additional cell lines examined. To confirm the results of the direct sequencing, cell line and normal adrenal DNA was subcloned and 10 clones for each cell line were sequenced. Consistent with the results of direct sequencing, CpG methylation was detected in cell lines but not in normal human adrenal DNA. However, there was no correlation between the extent CST6 CpG island methylation and upregulation of CST6 expression after 5-aza-dC treatment. We then proceeded to analyse CST6 CpG island methylation in 20 primary tumours (10 neuroblastomas and 10 phaeochromocytomas). None of the tumours showed significant promoter methylation.
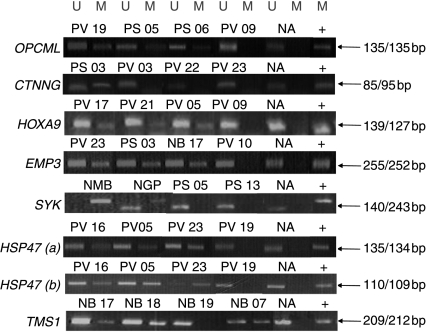
We then analysed the seven genes (OPCML, Junction plakoglobin (JUP), HOXA9, EMP3, SYK, HSP47 and TMS1) that a) had previously been shown to have be implicated in human tumourigenesis and b) demonstrated a >2-fold increase in expression after demethylation in at least one of three neuroblastoma cell lines. The promoter methylation status of OPCML, JUP, HOXA9, EMP3, SYK, HSP47 and TMS1 was analysed by previously published MSP primers in cell lines and in up to 33 phaeochromocytomas and 19 neuroblastomas (see Fig. 1 and Table 3). Three genes (HSP47, OPCML and HOXA9) demonstrated frequent (>10%) CpG island methylation in phaeochromocytoma tumours, but methylation of EMP3, although very frequent in neuroblastoma, was rare in phaeochromocytoma. Two CpG islands were examined for HSP47 methylation, one in the 5′ flanking region of HSP47 that encompasses a promoter and the second region around its transcriptional start site (Yang et al. 2007). In agreement with this previous study, similar patterns of methylation were seen in the two regions, although the promoter region showed a higher methylation frequency (90 and 52% in neuroblastoma and phaeochromocytoma tumours respectively; see Table 2). Methylation of EMP3 was relatively specific for neuroblastoma tumours (68 vs 6%, P=0.000006), whereas the frequency of HOXA9 promoter methylation was similar in the two tumour types. TMS1 and CTNNG displayed lower frequencies (<10%) of CpG island promoter methylation in phaeochromocytoma tumours and no promoter region methylation was detected in cell lines or primary tumours at SYK (Table 3).
Figure 1.
Methylation-specific PCR (MSP) of OPCML, CTNNG, HOXA9, EMP3, SYK, HSP47(a), HSP47(b) and TMS1 in neuroblastoma and phaeochromocytoma tumours. Bisulphite-modified DNA was amplified with primers specific for unmethylated (U) and methylated (M) DNA. Sample number above lane. NA, normal adrenal DNA. Sizes of the PCR products are indicated by arrows, U for unmethylated and M for methylated respectively. Positive control is Sss1-treated DNA.
Table 3.
Individual tumour–methylation patterns
| Gene | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Tumour type and number | OPCML | CTNNG | HOX9A | EMP3 | SYK | HSP47(A) | HSP47 (B) | TMS1 | MI |
| PV 03 | U | U | U | U | U | U | M | U | 0.125 |
| PV 04 | U | U | U | U | U | M | M | U | 0.25 |
| PV 05 | U | U | M | U | U | M | M | U | 0.375 |
| PV 06 | U | U | U | U | U | U | U | U | 0 |
| PV 07 | U | U | U | U | U | U | M | U | 0.125 |
| PV 08 | U | U | U | U | U | U | U | U | 0 |
| PV 09 | U | U | U | U | U | M | M | U | 0.25 |
| PV 10 | U | U | U | U | U | M | M | U | 0.25 |
| PV 11 | U | U | ND | U | ND | U | U | ND | 0 |
| PV 12 | U | U | ND | U | ND | M | M | ND | 0.25 |
| PV 13 | U | U | M | U | U | U | U | U | 0.125 |
| PV 15 | U | U | U | U | U | U | U | U | 0 |
| PV 16 | U | U | U | U | U | M | M | U | 0.25 |
| PV 17 | U | U | M | U | U | U | U | U | 0.125 |
| PV 18 | U | U | U | U | U | U | U | U | 0 |
| PV 19 | M | U | U | U | U | U | U | U | 0.125 |
| PV 21 | U | U | M | U | U | U | U | U | 0.125 |
| PV 22 | M | U | ND | U | ND | U | M | ND | 0.25 |
| PV 23 | U | U | U | M | U | M | M | U | 0.375 |
| PS 03 | U | M | M | M | U | M | M | M | 0.75 |
| PS 05 | M | U | U | U | U | U | U | U | 0.125 |
| PS 06 | M | U | U | U | U | M | M | U | 0.375 |
| PS 07 | U | U | U | U | U | U | U | M | 0.125 |
| PS 08 | U | U | U | U | U | U | U | U | 0 |
| PS 09 | U | U | U | U | U | U | M | U | 0.125 |
| PS 11 | U | U | ND | U | ND | U | U | ND | 0 |
| PS 12 | U | U | U | U | U | M | M | U | 0.25 |
| PS 13 | U | U | U | U | U | U | U | U | 0 |
| PS 14 | U | U | U | U | U | U | U | U | 0 |
| PS 15 | U | U | U | U | U | M | M | U | 0.25 |
| PS 16 | U | U | U | U | U | M | M | U | 0.25 |
| PS 17 | U | U | U | U | U | U | M | U | 0.125 |
| PS 18 | U | U | U | U | U | U | U | U | 0 |
| NB 01 | U | U | M | M | M | M | ND | 0.67 | |
| NB 03 | U | M | U | U | U | U | M | U | 0.25 |
| NB 04 | U | U | U | U | U | U | M | U | 0.125 |
| NB 05 | U | U | U | U | U | U | M | U | 0.125 |
| NB 06 | U | U | M | M | U | M | M | U | 0.5 |
| NB 07 | U | U | U | M | U | M | M | M | 0.375 |
| NB 08 | U | U | U | U | U | U | U | U | 0 |
| NB 09 | U | U | U | M | U | M | M | U | 0.375 |
| NB 10 | U | U | U | M | U | M | M | U | 0.375 |
| NB 11 | M | U | U | M | U | M | M | U | 0.5 |
| NB 12 | U | M | U | U | U | U | M | U | 0.25 |
| NB 13 | U | U | U | M | U | M | M | U | 0.375 |
| NB 14 | U | U | U | M | U | M | M | U | 0.375 |
| NB 15 | U | U | U | M | U | M | M | U | 0.375 |
| NB 16 | U | U | U | M | U | M | M | U | 0.375 |
| NB 17 | U | U | M | M | U | M | M | M | 0.625 |
| NB 18 | U | U | M | M | U | M | M | M | 0.625 |
| NB 19 | U | U | U | U | U | U | U | U | 0 |
| NB 20 | M | U | ND | M | ND | M | M | ND | 0.8 |
NB, neuroblastoma; PS, sporadic phaeochromocytoma; PV, VHL, associated phaeochromocytoma; U, unmethylated; M, methylated; ND, not determined (failed); MI, methylation index (number of methylated genes/number genes analysed).
Table 2.
Summary of gene-specific methylation data in VHL-associated and sporadic phaeochromocytomas and neuroblastoma tumours
| Phaeochromocytoma (% methylated) | ||||
|---|---|---|---|---|
| All | VHL | Sporadic | Neuroblastoma tumours (% methylated) | |
| OPCML | 12 (4/33) | 10.5 (2/19) | 14.3 (2/14) | 10.5 (2/19) |
| CTNNG | 3 (1/33) | 0 (0/19) | 7.1 (1/14) | 10.5 (2/19) |
| HOXA9 | 17.2 (5/29) | 25 (4/16) | 7.7 (1/13) | 22.2 (4/18) |
| EMP3 | 6.1 (2/33) | 5.3 (1/19) | 7.1 (1/14) | 68.4 (13/19) |
| SYK | 0 (0/29) | 0 (0/16) | 0 (0/13) | 0 (0/18) |
| HSP47 (A) | 36.4 (12/33) | 36.8 (7/19) | 35.7 (5/14) | 68.4 (13/19) |
| HSP47 (B) | 51.5 (17/33) | 52.6 (10/19) | 50 (7/14) | 89.5 (17/19) |
| TMS1 | 6.9 (2/29) | 0 (0/16) | 15.4 (2/13) | 16.7 (3/18) |
Data are percentages and numbers of tumour samples analysed. Methylation of EMP3 and HSP47 (A) and HSP47 (B) was significantly more frequent in neuroblastomas than phaeochromocytomas (P=0.001, P=0.02 and P=0.005 respectively).
No association was detected between methylation at OPCML, HOXA9 or HSP47 in individual tumours. There was also no association between methylation at OPCML or HSP47 and methylation as CASP 8 or HIC1 (Margetts et al. 2005). To analyse whether the different subtypes of neural crest tumours showed different levels of methylation, we combined data from the current study with additional data reported previously for FLIP, TSP1, DcR1, DcR2, DR4, DR5, CASP8 and HIC1 (Margetts et al. 2005) and estimated the mean percentage of loci methylated in each tumour sample. There were no significant differences in the mean percentage of loci methylated among neuroblastoma (mean 34.3%), VHL phaeochromocytoma (mean 26.8%) and sporadic phaeochromocytoma (mean 27.4%). We did not find any evidence that a particular gene was differentially methylated in VHL and sporadic phaeochromocytomas. To determine whether there was evidence for a subset of tumours with a CpG island methylator phenotype (CIMP), the distribution of number of methylated loci per tumour was compared with that expected from a Poisson distribution. However, there were no significant differences between the observed and expected distributions (P=0.3751).
Discussion
Phaeochromocytoma and neuroblastoma are both derived from the neural crest and there are overlaps between the regions of allele loss and patterns of TSG methylation observed in the two tumour types. In the absence of a human phaeochromocytoma cell line, we analysed gene expression changes following treatment with a demethylating agent in three neuroblastoma cell lines. Such functional epigenomic screens have proven to be a successful strategy to identify epigenetically inactivated TSGs in a number of different tumour types including oesophageal, pancreatic and prostate (Yamashita et al. 2002, Sato et al. 2003, Lodygin et al. 2005). In addition, using this strategy in RCC cell lines, we identified HAI-2/SPINT2 as a novel epigenetically inactivated RCC TSG (Morris et al. 2005).
Although our selection criteria for investigating ‘candidate epigenetically silenced TSGs’ would be expected to result in some false negative prediction, the major problem we encountered was the absence of promoter region methylation in many of the genes we analysed. In addition, two genes that did show evidence of promoter methylation in neuroblastoma cell lines did not prove to be methylated in primary tumours. Whilst these results were disappointing, they are not unique to neuroblastoma cell lines. Thus, in a similar study of four RCC cell lines, we identified SPINT2 as a novel epigenetically inactivated renal TSG. However, analysis of a further 60 genes that were differentially expressed after demethylation revealed that only six (four of which had been reported previously) demonstrated primary tumour-specific promoter methylation (Morris et al. 2008). Thus, this approach has a relatively low specificity for identifying methylated TSGs. Analysis of a larger number of genes might have led to the identification of novel TSGs, but reducing the stringency of the selection criteria would also probably further lower specificity. A more effective plan of investigation might be to combine ‘functional epigenomics’ experiments with strategies to directly identify methylated DNA (e.g. MeDIP), as this should help exclude genes whose expression is upregulated after demethylation but do not demonstrate promoter methylation (Wilson et al. 2006). Nevertheless, it is possible that candidate TSGs that are unmethylated but upregulated by demethylation may prove to be downstream of epigenetically inactivated TSGs. Thus, we used the Oncomine (http://www.oncomine.org/) data analysis tool to interrogate gene expression data reported by Asgharzadeh et al. (2006) for 19 genes that were upregulated by demethylation but did not show detectable CpG methylation (SNN, RASD1, ARPC4, PTK2B, TFAP4, ZFP36L2, ARHGDIA, suppressor of lin-12-like (SEL1L), LTBP3, smoothelin (SMTH), ST3 β-galactoside α-2,3-sialyltransferase 2 (ST3GAL2), v-rel reticuloendotheliosis viral oncogene homologue A (RELA), tripartite motif-containing 3 (TRIM3), chromodomain helicase DNA -binding protein 5 (CHD5), PRKA2R, AK1, UCP2, NUP62, SLC2A4). Out of 19 genes, 4 (adenylate kinase 1; AK1, TFAP4, SNN and SEL1L) were differentially expressed levels between relapsing and non-relapsing tumours. In addition, CHD5 has been identified as a candidate 1p36 neuroblastoma TSG (Bagchi et al. 2007).
There was a higher frequency of promoter methylation in primary tumours among the seven genes that had been previously implicated in human tumourigenesis and were selected according to the less strict criteria (upregulated in one or more cell lines). Three of these genes, HSP47, HOXA9 and OPCML were methylated in >10% of phaeochromocytomas.
Both heat-shock protein 47 (HSP47) and OPCML map to 11q (11q13.5 and 11q25 respectively) and chromosome 11q allele loss is frequent in both tumour types (Yokogoshi et al. 1990, Sun et al. 2006, George et al. 2007). HSP47 encodes a collagen-specific molecular chaperone and is essential for the production and maturation of collagens I and IV (Sauk et al. 2005). Type I collagen negatively regulates cell proliferation and is deficient in aggressive neuroblastoma tumours (Yang et al. 2004). Previously, expression of HSP47 was reported to be upregulated after treatment with 5-aza-dC in the neuroblastoma cell line NB2-W-S and promoter region methylation was detected in neuroblastoma cell lines and 4/7 primary neuroblastoma tumours analysed (Yang et al. 2004). In addition, expression of HSP47 correlated with that of collagen I and IV. We have confirmed and extended the analysis of HSP47 promoter methylation in neuroblastoma and demonstrated that the HSP47 promoter is methylated in ∼50% of phaeochromocytomas.
HOXA9 promoter region methylation was detected in about one-fifth of neuroblastoma and phaeochromocytomas. Several Hox genes have been reported to be methylated in human cancers, e.g. HOXB13 and HOXA5, in renal and breast cancer respectively (Okuda et al. 2006, Piotrowski et al. 2006). HOXA9 promoter methylation was identified previously in lung cancer (Rauch et al. 2007) and in neuroblastoma (Alaminos et al. 2004), but not in phaeochromocytoma. Previously, we found significant differences in the frequency of HIC1 and CASP8 methylation between VHL-associated and sporadic phaeochromocytomas, but in this study we did not identify any further differentially methylated genes.
OPCML (OPCML/cell adhesion molecule-like) encodes a member of the IgLON subfamily in the immunoglobulin protein superfamily and was identified as a candidate TSG following the identification of extensive promoter methylation in ovarian cancer (Sellar et al. 2003). Ectopic expression of the OPCML gene product suppressed cell growth in vitro and tumour growth in vivo in nude mice models. Promoter methylation associated silencing of OPCML has also been demonstrated in hepatocellular carcinomas (Liu et al. 2006) and gliomas and other brain tumours (including medulloblastomas) (Reed et al. 2007). To our knowledge, OPCML promoter methylation has not previously been reported in neuroblastoma or phaeochromocytoma.
EMP3 is a peripheral myelin protein involved in cell proliferation and cell–cell interactions. Previously EMP3 expression was reported to be downregulated in glioma and neuroblastoma tumours, and EMP3 promoter methylation was detected in 24% of neuroblastomas (Alaminos et al. 2005). However, although we confirmed frequent EMP3 promoter methylation in neuroblastoma, EMP3 methylation did not appear to make a significant contribution to phaeochromocytoma tumourigenesis.
Infrequent, or absent, promoter methylation was found for CTNNG (γ-catenin, also known as JUP) and SYK in phaeochromocytoma. Although Amitay et al. (2001) reported reduced expression of CTNNG in 9/20 neuroblastoma tumours, we observed promoter methylation in only 11% of neuroblastoma tumours and did not detect promoter methylation in the cell lines that demonstrated an increase expression of CTNNG after 5-aza-dC treatment. This suggests that promoter methylation of CTNNG does not play a large role in its regulation of expression in neural crest tumours. SYK (spleen tyrosine kinase) and TMS1 (also known as ASC, apoptosis speck-like protein containing CARD) have been reported to be methylated in 60 and 80% of neuroblastoma cell lines respectively (Alaminos et al. 2004), our results suggest that methylation is less frequent in neuroblastoma primary tumours and is infrequent (<10%) in phaeochromocytoma. The CIMP methylator phenotype is well described in a subset of colorectal cancers (Weisenberger et al. 2006). However, we did not find evidence of a CIMP subgroup in neural crest tumours and mean methylation index did not differ between neuroblastomas, VHL and sporadic phaeochromoctromas. Our findings demonstrate that many genes that are methylated in neuroblastoma are also methylated in phaeochromocytoma. Identification of methylated genes in neural crest tumours will provide potential biomarkers and can provide insights into the molecular pathogenesis of tumour development in these disorders.
Declaration of interest
The authors declare that there is no conflict of interest that would prejudice the impartiality of this scientific work.
Funding
We thank the Cancer Research UK and the University of Birmingham for financial support.
References
- Alaminos M, Davalos V, Cheung NK, Gerald WL, Esteller M. Clustering of gene hypermethylation associated with clinical risk groups in neuroblastoma. Journal of the National Cancer Institute. 2004;96:1208–1219. doi: 10.1093/jnci/djh224. [DOI] [PubMed] [Google Scholar]
- Alaminos M, Davalos V, Ropero S, Setien F, Paz MF, Herranz M, Fraga MF, Mora J, Cheung NK, Gerald WL, et al. EMP3, a myelin-related gene located in the critical 19q13.3 region, is epigenetically silenced and exhibits features of a candidate tumor suppressor in glioma and neuroblastoma. Cancer Research. 2005;65:2565–2571. doi: 10.1158/0008-5472.CAN-04-4283. [DOI] [PubMed] [Google Scholar]
- Amitay R, Nass D, Meitar D, Goldberg I, Davidson B, Trakhtenbrot L, Brok-Simoni F, Ben-Ze'ev A, Rechavi G, Kaufmann Y. Reduced expression of plakoglobin correlates with adverse outcome in patients with neuroblastoma. American Journal of Pathology. 2001;159:43–49. doi: 10.1016/S0002-9440(10)61671-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asgharzadeh S, Pique-Regi R, Sposto R, Wang H, Yang Y, Shimada H, Matthay K, Buckley J, Ortega A, Seeger RC. Prognostic significance of gene expression profiles of metastatic neuroblastomas lacking MYCN gene amplification. Journal of the National Cancer Institute. 2006;98:1193–1203. doi: 10.1093/jnci/djj330. [DOI] [PubMed] [Google Scholar]
- Astuti D, Latif F, Dallol A, Dahia PL, Douglas F, George E, Skoldberg F, Husebye ES, Eng C, Maher ER. Gene mutations in the succinate dehydrogenase subunit SDHB cause susceptibility to familial pheochromocytoma and to familial paraganglioma. American Journal of Human Genetics. 2001a;69:49–54. doi: 10.1086/321282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astuti D, Douglas F, Lennard TW, Aligianis IA, Woodward ER, Evans DG, Eng C, Latif F, Maher ER. Germline SDHD mutation in familial phaeochromocytoma. Lancet. 2001b;357:1181–1182. doi: 10.1016/S0140-6736(00)04378-6. [DOI] [PubMed] [Google Scholar]
- Astuti D, Agathanggelou A, Honorio S, Dallol A, Martinsson T, Kogner P, Cummins C, Neumann HP, Voutilainen R, Dahia P, et al. RASSF1A promoter region CpG island hypermethylation in phaeochromocytomas and neuroblastoma tumours. Oncogene. 2001c;20:7573–7577. doi: 10.1038/sj.onc.1204968. [DOI] [PubMed] [Google Scholar]
- Astuti D, Hart-Holden N, Latif F, Lalloo F, Black GC, Lim C, Moran A, Grossman AB, Hodgson SV, Freemont A, et al. Genetic analysis of mitochondrial complex II subunits SDHD, SDHB and SDHC in paraganglioma and phaeochromocytoma susceptibility. Journal of Clinical Endocrinology. 2003;59:728–733. doi: 10.1046/j.1365-2265.2003.01914.x. [DOI] [PubMed] [Google Scholar]
- Breault JE, Shiina H, Igawa M, Ribeiro-Filho LA, Deguchi M, Enokida H, Urakami S, Terashima M, Nakagawa M, Kane CJ, Carroll PR, Dahiya R. Methylation of the gamma-catenin gene is associated with poor prognosis of renal cell carcinoma. Clinical Cancer Research. 2005;11:557–564. [PubMed] [Google Scholar]
- Chrisoulidou A, Kaltsas G, Ilias I, Grossman AB. The diagnosis and management of malignant phaeochromocytoma and paraganglioma. Endocrine-Related Cancer. 2007;14:569–585. doi: 10.1677/ERC-07-0074. [DOI] [PubMed] [Google Scholar]
- Conway KE, McConnell BB, Bowring CE, Donald CD, Warren ST, Vertino PM. TMS1, a novel proapoptotic caspase recruitment domain protein, is a target of methylation-induced gene silencing in human breast cancers. Cancer Research. 2000;60:6236–6242. [PubMed] [Google Scholar]
- Disick GI, Palese MA. Extra-adrenal pheochromocytoma: diagnosis and management. Current Urology Reports. 2007;8:83–88. doi: 10.1007/s11934-007-0025-5. [DOI] [PubMed] [Google Scholar]
- Eng C, Crossey PA, Mulligan LM, Healey CS, Houghton C, Prowse A, Chew SL, Dahia PL, O'Riordan JL, Toledo SP. Mutations in the RET proto-oncogene and the von Hippel–Lindau disease tumour suppressor gene in sporadic and syndromic phaeochromocytomas. Journal of Medical Genetics. 1995;32:934–937. doi: 10.1136/jmg.32.12.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George RE, Attiyeh EF, Li S, Moreau LA, Neuberg D, Li C, Fox EA, Meyerson M, Diller L, Fortina P, et al. Genome-wide analysis of neuroblastomas using high-density single nucleotide polymorphism arrays. PLoS ONE. 2007;2:e255. doi: 10.1371/journal.pone.0000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Roqueplo AP. New advances in the genetics of pheochromocytoma and paraganglioma syndromes. Annals of the New York Academy of Sciences. 2006;1073:112–121. doi: 10.1196/annals.1353.012. [DOI] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. PNAS. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofstra RM, Cheng NC, Hansen C, Stulp RP, Stelwagen T, Clausen N, Tommerup N, Caron H, Westerveld A, Versteeg R, et al. No mutations found by RET mutation scanning in sporadic and hereditary neuroblastoma. Human Genetics. 1996;97:362–364. doi: 10.1007/BF02185773. [DOI] [PubMed] [Google Scholar]
- Lind GE, Skotheim RI, Fraga MF, Abeler VM, Esteller M, Lothe RA. Novel epigenetically deregulated genes in testicular cancer include homeobox genes and SCGB3A1 (HIN-1) Journal of Pathology. 2006;210:441–449. doi: 10.1002/path.2064. [DOI] [PubMed] [Google Scholar]
- Liu WJ, Wang L, Wang JP, Li JQ, Zhang CO, Zheng L, Yuan YF. Correlations of CpG island methylator phenotype and OPCML gene methylation to carcinogenesis of hepatocellular carcinoma. Ai Zheng. 2006;25:696–700. [PubMed] [Google Scholar]
- Lodygin D, Epanchintsev A, Menssen A, Diebold J, Hermeking H. Functional epigenomics identifies genes frequently silenced in prostate cancer. Cancer Research. 2005;65:4218–4227. doi: 10.1158/0008-5472.CAN-04-4407. [DOI] [PubMed] [Google Scholar]
- Margetts CDE, Astuti D, Gentle DC, Cooper WN, Cascon A, Catchpoole D, Robledo M, Neumann HPH, Latif F, Maher ER. Epigenetic analysis of HIC1, CASP8, FLIP, TSP1, DCR1, DCR2, DR4, DR5, KvDMR1, H19 and preferential 11p15.5 maternal-allele loss in von Hippel–Lindau and sporadic phaeochromocytomas. Endocrine-Related Cancer. 2005;12:161–172. doi: 10.1677/erc.1.00865. [DOI] [PubMed] [Google Scholar]
- Morris MR, Gentle DC, Abdulrahman M, Maina EN, Gupta K, Banks RE, Wiesener MS, Kishida T, Yao M, The B, et al. Tumour suppressor activity and epigenetic inactivation of hepatocyte growth factor activator inhibitor type 2/SPINT2 in papillary and clear cell renal cell carcinoma. Cancer Research. 2005;65:4598–4606. doi: 10.1158/0008-5472.CAN-04-3371. [DOI] [PubMed] [Google Scholar]
- Morris MR, Gentle D, Abdulrahman M, Clarke N, Brown M, Kishida T, Yao M, The BT, Latif F, Maher ER. Functional epigenomics approach to identify methylated candidate tumour suppressor genes in renal cell carcinoma. British Journal of Cancer. 2008;98:496–501. doi: 10.1038/sj.bjc.6604180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann HP, Bausch B, McWhinney SR, Bender BU, Gimm O, Franke G, Schipper J, Klisch J, Altehoefer C, Zerres K, et al. Germ-line mutations in nonsyndromic pheochromocytoma. New England Journal of Medicine. 2002;346:1459–1466. doi: 10.1056/NEJMoa020152. [DOI] [PubMed] [Google Scholar]
- Okuda H, Toyota M, Ishida W, Furihata M, Tsuchiya M, Kamada M, Tokino T, Shuin T. Epigenetic inactivation of the candidate tumor suppressor gene HOXB13 in human renal cell carcinoma. Oncogene. 2006;25:1733–1742. doi: 10.1038/sj.onc.1209200. [DOI] [PubMed] [Google Scholar]
- Piotrowski A, Benetkiewicz M, Menzel U, de Ståhl TD, Mantripragada K, Grigelionis G, Buckley P, Jankowski M, Hoffman J, Bała D, et al. Microarray-based survey of CpG islands identifies concurrent hyper- and hypomethylation patterns in tissues derived from patients with breast cancer. Genes, Chromosomes and Cancer. 2006;45:656–667. doi: 10.1002/gcc.20331. [DOI] [PubMed] [Google Scholar]
- Rauch T, Wang Z, Zhang X, Zhong X, Wu X, Lau SK, Kernstine KH, Riggs AD, Pfeifer GP. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. PNAS. 2007;104:5527–5532. doi: 10.1073/pnas.0701059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JE, Dunn JR, Du Plessis DG, Shaw EJ, Reeves P, Gee AL, Warnke PC, Sellar GC, Moss DJ, Walker C. Expression of cellular adhesion molecule ‘OPCML’ is down-regulated in gliomas and other brain tumours. Neuropathology and Applied Neurobiology. 2007;33:77–85. doi: 10.1111/j.1365-2990.2006.00786.x. [DOI] [PubMed] [Google Scholar]
- Sato N, Fukushima N, Maitra A, Matsubayashi H, Yeo CJ, Cameron JL, Hruban RH, Goggins M. Discovery of novel targets for aberrant methylation in pancreatic carcinoma using high-throughput microarrays. Cancer Research. 2003;63:3735–3742. [PubMed] [Google Scholar]
- Sauk JJ, Nikitakis N, Slavash H. Hsp47 a novel collagen binding serpin chaperone, autoantigen and therapeutic target. Frontiers in Bioscience. 2005;10:107–118. doi: 10.2741/1513. [DOI] [PubMed] [Google Scholar]
- Sellar GC, Watt KP, Rabiasz GJ, Stronach EA, Li L, Miller EP, Massie CE, Miller J, Contreras-Moreira B, Scott D, et al. OPCML at 11q25 is epigenetically inactivated and has tumor-suppressor function in epithelial ovarian cancer. Nature Genetics. 2003;34:337–343. doi: 10.1038/ng1183. [DOI] [PubMed] [Google Scholar]
- Sun HY, Cui B, Su DW, Jin XL, Sun FK, Zu Y, Jiang L, Wang WQ, Ning G. LOH on chromosome 11q, but not SDHD and Men1 mutations was frequently detectable in Chinese patients with pheochromocytoma and paraganglioma. Endocrine. 2006;30:307–312. doi: 10.1007/s12020-006-0009-0. [DOI] [PubMed] [Google Scholar]
- Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nature Genetics. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- Wilson I, Davies J, Weber M, Brown C, Alvarez CE, MacAulay C, Schübeler D, Lam WL. Epigenomics: mapping the methylome. Cell Cycle. 2006;5:155–158. doi: 10.4161/cc.5.2.2367. [DOI] [PubMed] [Google Scholar]
- Yamashita K, Upadhyay S, Osada M, Hoque MO, Xiao Y, Mori M, Sato F, Meltzer SJ, Sidransky D. Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. Cancer Cell. 2002;2:485–495. doi: 10.1016/s1535-6108(02)00215-5. [DOI] [PubMed] [Google Scholar]
- Yang Q, Liu S, Tian Y, Hasan C, Kersey D, Salwen HR, Chlenski A, Perlman EJ, Cohn SL. Methylation-associated silencing of the heat shock protein 47 gene in human neuroblastoma. Cancer Research. 2004;64:4531–4538. doi: 10.1158/0008-5472.CAN-04-0956. [DOI] [PubMed] [Google Scholar]
- Yang Q, Kiernan CM, Tian Y, Salwen HR, Chlenski A, Brumback BA, London WB, Cohn SL. Methylation of CASP8, DCR2, and HIN-1 in neuroblastoma is associated with poor outcome. Clinical Cancer Research. 2007;13:3191–3197. doi: 10.1158/1078-0432.CCR-06-2846. [DOI] [PubMed] [Google Scholar]
- Yokogoshi Y, Yoshimoto K, Saito S. Loss of heterozygosity on chromosomes 1 and 11 in sporadic pheochromocytomas. Japanese Journal of Cancer Research. 1990;81:632–638. doi: 10.1111/j.1349-7006.1990.tb02620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]