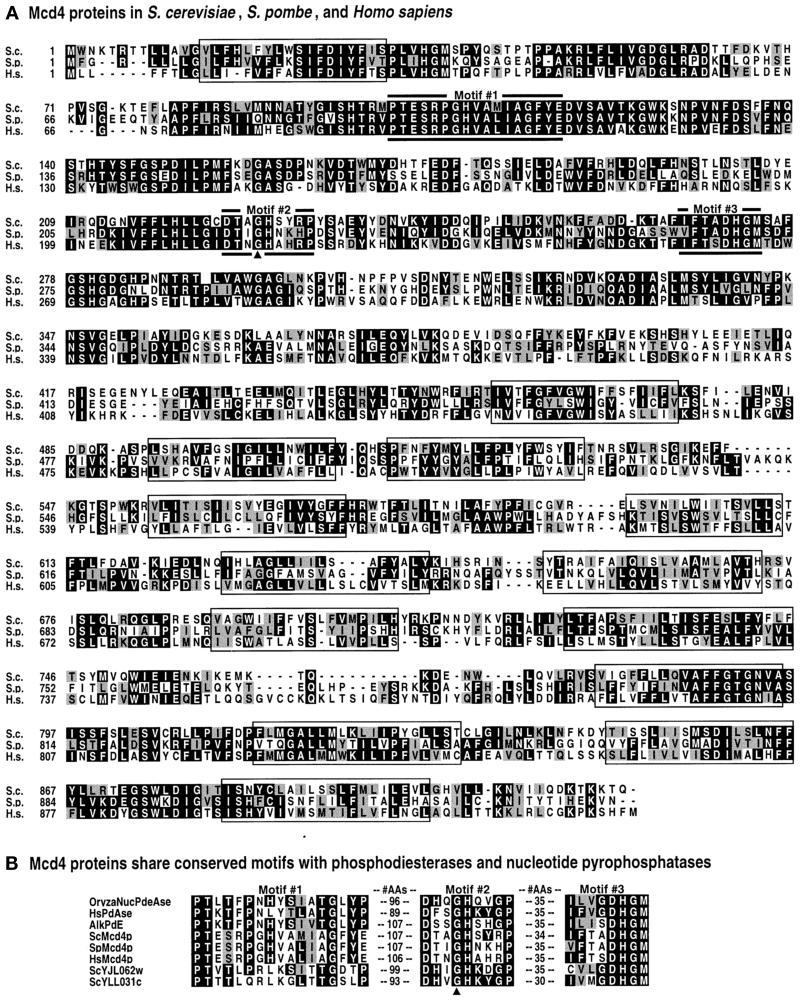
Figure 2.
Mcd4 protein sequence alignments and motifs. (A) Alignment of Mcd4 proteins from S. cerevisiae, S. pombe, and Homo sapiens, using the CLUSTALW Multiple Sequence Aligment Program. Regions of amino acid identity are shaded black. Regions of amino acid identity are shaded gray. TMDs are in boxes. Motifs 1, 2, and 3 (labeled; also see part B) are denoted by thick lines. (B) Three conserved motifs found in phosphodiesterases, pyrophosphatases, Mcd4p, and Mcd4p homologues. Motifs were identified using MEME and MAST analyses. Shading is used to highlight identities and similarities between the known enzymes and Mcd4p and/or its homologues: within a given amino acid position, the most prevalent residue in Mcd4p or its homologues that is identical to an amino acid in the same position in a known enzyme are shaded black, while those that are similar are shaded gray. Amino acids that are identical or similar only between Mcd4p and its homologues are not shaded. “AAs” signifies the number of amino acids separating the motifs. In both panels A and B, the arrowhead points to glycine 227, which is mutated to glutamic acid in S. cerevisiae mcd4–174.