Abstract
Transfer of class 1 integron-mediated antibiotic resistance genes has been demonstrated under laboratory conditions. However, there is no information concerning the transfer of these genes in an agricultural environment. The present study sought to determine if integron-mediated streptomycin and sulfisoxazole resistance genes could be transferred from Shiga toxin-producing Escherichia coli (STEC) strains 6-20 (O157:H7) and 7-63 (O111:H8) to the susceptible strain E. coli K-12 MG1655 in bovine feces (pH 5.5, 6.0, or 6.5) and storm water (pH 5, 6, 7, or 8) at 4, 15, and 28°C, which are average seasonal temperatures for winter, spring-fall, and summer, respectively, in the Griffin, GA, area. The results indicated that at 28°C, the integron-mediated antibiotic resistance genes were transferred from both of the STEC donors in bovine feces. Higher conjugation efficiencies were, however, observed in the conjugation experiments involving STEC strain 6-20. In storm water, the resistance genes were transferred only from STEC strain 6-20. Greater numbers of transconjugants were recovered in the conjugation experiments performed with pH 6.5 bovine feces and with pH 7 storm water. Antibiotic susceptibility tests confirmed the transfer of integron-mediated streptomycin resistance and sulfisoxazole resistance, as well as the transfer of non-integron-mediated oxytetracycline resistance and tetracycline resistance in the transconjugant cells. These results suggest that the antibiotic resistance genes in STEC could serve as a source of antibiotic resistance genes disseminated via conjugation to susceptible cells of other E. coli strains in an agricultural environment.
Shiga toxin-producing Escherichia coli (STEC) has been associated with food-borne outbreaks of infection worldwide. The diseases caused by STEC range from mild diarrhea to very severe conditions, such as hemolytic-uremic syndrome. The organisms responsible for these diseases have been traced to farm animals, primarily cattle (3, 8). They have been isolated from manure, water troughs, feedlots, and other places on cattle farms (8). The occurrence of STEC on animal carcasses and retail meats corresponds to the level of STEC contamination at the farm level (1). Human STEC infections associated with the consumption of contaminated fresh produce and water have been linked to the organisms originating from such farm areas (10; http://www.fda.gov/bbs/topics/NEWS/2007/NEW01593.html).
Antibiotic use in the agricultural environment is believed to play a key role in dissemination of antibiotic resistance genes among bacteria (14, 15, 20). Cells of bacteria acquire antibiotic resistance through genetic mutation and horizontal transfer of antibiotic resistance genes in response to selective pressures. The increased prevalence of antibiotic resistance among pathogens such as STEC has raised concerns because some of the antibiotics are used in treatment of infectious diseases in both humans and animals (13, 24). It has been suggested that overuse of antibiotics in animal husbandry creates a threat to human and veterinary medicine.
Antibiotic resistance, including resistance to streptomycin, sulfonamides, and tetracycline, has been observed primarily among clinical isolates of STEC (26). The genes for resistance to antibiotics can be transmitted through conjugative plasmids to homologous and heterologous bacterial species under laboratory conditions (19, 24, 28). The transfer of antibiotic resistance genes is often related to integrons, which are capable of capturing and inserting antibiotic resistance genes into their structure (7). The transfer of integron-mediated antibiotic resistance genes has often been studied in controlled laboratory conditions; however, no information concerning the transfer of such genes under simulated agricultural conditions has been available previously.
The goal of the present study was to determine whether the transfer of integron-mediated antibiotic resistance genes from STEC to generic E. coli could take place in samples of materials commonly found in the farm environment, such as storm water and bovine feces, and whether environmental temperature and sample pH could influence the frequency of transfer of the antibiotic resistance genes. It has been reported that the average seasonal temperatures for winter, spring-fall, and summer in the Griffin, GA, area are 4, 15 and 28°C, respectively (http://www.weather.com). The average pH values of natural water range from 5 in acid lakes to 8 in estuaries (22, 27), while the fecal material of cattle has average pH values ranging from 5.5 to 6.5 (9; http://www.livestocktrail.uiuc.edu/dairynet/paperDisplay.cfm?ContentID=648).
MATERIALS AND METHODS
Bacterial strains and environmental samples.
Two STEC isolates of bovine origin, 6-20 (O157:H7) and 7-63 (O111:H8), both from our laboratory collection, were used as donor strains in this study. These isolates carry class 1 integrons which are responsible for their resistance to streptomycin and sulfisoxazole (S. Nagachinta and J. Chen, unpublished data). They are also resistant to novobiocin, tetracycline, and oxytetracycline, characteristics which are likely mediated by an antibiotic resistance plasmid. The recipient strain was a nalidixic acid-resistant derivative of E. coli K-12 strain MG1655 CA32 provided by Wondwossen A. Gebreyes at North Carolina State University, Raleigh. The cultures were grown on MacConkey agar or sorbitol MacConkey (SMAC) (Becton, Dickinson and Co., Sparks, MD) at 37°C for 16 h. A single colony of each culture was inoculated onto tryptic soy agar (TSA) (Becton, Dickinson and Co., Sparks, MD) and incubated under the conditions described above.
Storm water was collected from storm water discharge in Griffin, GA (pH at the time of collection, 6.4). The collected storm water was dispensed into glass bottles (40 ml each) before it was autoclaved at 121°C for 15 min. Bovine feces were collected from a single cattle farm in Griffin, GA (pH at the time of collection, 6.5). The sample was distributed into glass beakers (40 g in each beaker) and autoclaved at 121°C for 30 min. The sterility of the storm water and bovine feces was confirmed by plating the autoclaved samples on TSA plates to determine bacterial growth.
Preparation of donor and recipient cells.
Single colonies of the donor and recipient strains on TSA plates were transferred into tryptic soy broth (TSB), and the inoculated broth cultures were incubated at 37°C for 16 h. The resulting cultures (108 CFU/ml) were then centrifuged at 3,000 × g for 15 min. The pellet obtained from each culture was resuspended in a sterile saline solution (0.85% NaCl) in order to remove the nutrient residues from the culture medium.
Conjugation in storm water.
Storm water (40 ml) was inoculated with donor and recipient cells prepared as described above. The ratio of donor cells to recipient cells used for inoculation was 0.4:4 ml (108 CFU/ml). The inoculated storm water samples were held for 2 weeks in incubation chambers with ambient temperatures of 4, 15, and 28°C, which are the average ambient winter, fall-spring, and summer temperatures in the Griffin, GA, area, respectively. These average temperatures were calculated based on the average high and low monthly temperatures of the area obtained at the website http://weather.msn.com/monthly_averages.aspx?&wealocations=wc%3aUSGA0251&setunit=C. Storm water inoculated with only the donor cells (0.4 ml) or only the recipient cells (4 ml), both at a concentration of 108 CFU/ml, and uninoculated storm water samples were included in the conjugation experiments as negative controls. The numbers of transconjugant cells were determined every 2 days during the experimental period, and the experiment was replicated in two separate trials.
Subsequently, the effect of storm water pH on the efficiency of conjugation was investigated. The response surface methodology was used to design the experiments. The pH of the storm water was adjusted, with either HCl or NaOH, to 5, 6, 7, and 8, which represented the pH values of natural water sources, such as lake and estuary waters (22, 27). The storm water samples were held at 28°C for 1 week. Samples were taken every 2 days, and the numbers of antibiotic-resistant transconjugant cells were determined.
Conjugation in bovine feces.
A similar design was used in the conjugation experiment performed with bovine feces. The prepared donor and recipient cells (108 CFU/ml) were inoculated into 40 g of sterile fecal material using the same ratio and volumes of donor and recipient cells that were used in the storm water experiment. The inoculated feces were mixed thoroughly using sterile, disposable, plastic pipette tips. Uninoculated fecal samples and samples inoculated with only the donor and recipient cells were included in the study as negative controls. All samples were held for 2 weeks at the temperatures indicated above. Similar to the conjugation experiment performed with storm water, the numbers of transconjugant cells in bovine feces were determined every 2 days during the incubation period, and the experiment was replicated in two separate trials.
To study the influence of pH on the conjugation efficiency in bovine feces, the pH of the fecal material was adjusted with either HCl or NaOH to 5.5, 6.0, and 6.5, which represent the pH range for bovine feces (9; http://www.livestocktrail.uiuc.edu/dairynet/paperDisplay.cfm?ContentID=648). The donor and recipient cells were inoculated into the fecal material at the ratio indicated above. The inoculated samples were mixed well and kept at 28°C for 1 week. The numbers of antibiotic-resistant transconjugants were determined every 2 days.
Estimation of conjugation efficiency.
To determine the numbers of transconjugants derived from the conjugation conducted with storm water, 1 ml of each inoculated storm water sample was placed into a disposable, conical polypropylene centrifuge tube and centrifuged at 3,000 × g for 15 min (GS-6R centrifuge; Beckman, Fullerton, CA). The cell pellets were resuspended in 100 μl of a sterile saline solution before they were plated onto SMAC supplemented with nalidixic acid (100 μg/ml) and one of the following antibiotics: streptomycin (100 μg/ml), tetracycline (30 μg/ml), or oxytetracycline (30 μg/ml). The sizes of the populations of the recipient cells (per ml of culture) were determined by plating appropriate dilutions of each E. coli MG1655 culture onto SMAC plates supplemented with only nalidixic acid (100 μg/ml).
To determine the number of transconjugant cells in bovine feces, 1 g of the fecal material was suspended in 1 ml of a sterile saline solution in a conical tube as described above and centrifuged at 500 × g for 10 min in order to remove the solid substances in the fecal material. The supernatant was then collected and recentrifuged at 3,000 × g for 15 min to sediment the E. coli cells. The cell pellet was resuspended in 100 μl of a sterile saline solution and inoculated onto selective agar plates as described above. When necessary, the supernatants were serially diluted and appropriate dilutions were inoculated onto the selective agar plates.
The absence of antibiotic-resistant transconjugant cells in the negative control storm water and bovine fecal samples was confirmed by plating the samples on selective plates as described above. The conjugation efficiency was calculated by determining the ratio of the number of transconjugant cells to the number of recipient cells per ml of storm water or per g of bovine feces.
Analysis of transconjugants.
It was shown in a previous study conducted in our laboratory that resistance to tetracycline and resistance to oxytetracycline, which are mediated by an antibiotic resistance plasmid, were cotransferred with resistance to streptomycin, a trait mediated by a class 1 integron. Therefore, the antibiotic-resistant transconjugant cells derived from each conjugation experiment in the present study were tested to determine their resistance to the antibiotics that were not used as the selective markers in the conjugation experiment. The presence of the class 1 integrons in the transconjugant cells was confirmed by determination of their susceptibility to streptomycin and sulfisoxazole. A standard disk diffusion assay was performed according to the guidelines provided by the NCCLS (currently the Clinical and Laboratory Standards Institute) (17). Selected transconjugant cells obtained from each selective plate were inoculated onto a TSA plate to obtain a single colony and incubated at 37°C for 12 h. A single colony of each culture was transferred into TSB (Becton, Dickinson and Co., Sparks, MD) and incubated under the same conditions. The concentrations of the cell populations in the resulting cultures were adjusted with TSB so that the optical density at 600 nm was 0.1 (Novaspec II visible spectrophotometer; Pharmacia Biotech, Cambridge, United Kingdom). The adjusted cell cultures were inoculated onto Mueller-Hinton agar (Becton, Dickinson and Co., Sparks, MD) plates with sterile cotton swabs and allowed to dry for 5 min before application of antibiotic disks (Sensi-Disc antimicrobial susceptibility test disks; BBL, Becton Dickinson) onto the plates using sterile forceps. The plates were incubated at 37°C for 16 h. The diameter of the zone of growth inhibition around each disk was determined to the nearest whole millimeter, and each cell culture tested was determined to be resistant, intermediate, or susceptible to the antibiotics used according to the guidelines provided by the supplier of the antibiotic disks.
RESULTS
Influence of temperature.
Among the three temperatures used in this study, antibiotic-resistant transconjugants were recovered only from the storm water and bovine feces held at 28°C (Table 1). The antibiotic resistance genes were transferred at higher frequencies from STEC 6-20 than from 7-63 at this temperature. In bovine feces, the frequency of transfer of antibiotic resistance genes from STEC 6-20 was approximately 100 times higher than the frequency of transfer of antibiotic resistance genes from STEC 7-63 (Table 1). In storm water, however, only the antibiotic resistance genes from 6-20 were transferred.
TABLE 1.
Frequencies of antibiotic resistance gene transfer from STEC isolates 6-20 and 7-63 to E. coli K-12 recipient strain MG1655 in storm water and bovine feces held at 28°C
| Donor strain | Selective antibiotic | Transfer frequencies at different sampling timesa
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Day 1 | Day 3 | Day 5 | Day 7 | Day 9 | Day 11 | Day 13 | Avg | ||
| Storm water | |||||||||
| 6-20 | Oxytetracycline | 8.93 × 10−7 | 1.86 × 10−6 | 7.50 × 10−7 | 1.04 × 10−6 | 8.93 × 10−7 | 9.64 × 10−7 | 1.04 × 10−6 | 1.06 × 10−6 |
| 6-20 | Tetracycline | <1 × 10−8 | <1 × 10−8 | <1 × 10−8 | 2.50 × 10−7 | <1 × 10−8 | <1 × 10−8 | <1 × 10−8 | <4.43 × 10−8 |
| Bovine feces | |||||||||
| 6-20 | Oxytetracycline | 3.75 × 10−6 | 7.65 × 10−6 | 7.10 × 10−6 | 1.26 × 10−5 | 7.25 × 10−6 | 1.12 × 10−5 | 1.71 × 10−5 | 9.52 × 10−6 |
| 6-20 | Tetracycline | 1.59 × 10−6 | 1.32 × 10−6 | 1.22 × 10−6 | 3.04 × 10−6 | 8.12 × 10−6 | 2.29 × 10−5 | 2.96 × 10−5 | 9.68 × 10−6 |
| 7-63 | Oxytetracycline | 1.01 × 10−7 | 1.01 × 10−7 | 2.90 × 10−8 | 1.45 × 10−8 | 4.35 × 10−8 | <1 × 10−8 | 1.45 × 10−8 | <4.45 × 10−8 |
| 7-63 | Tetracycline | <1 × 10−8 | <1 × 10−8 | <1 × 10−8 | <1 × 10−8 | 4.35 × 10−8 | 4.35 × 10−8 | 2.90 × 10−8 | <2.23 × 10−8 |
The detection limit was 1 × 10−8.
In storm water held at 28°C, the average transfer frequencies showed that the numbers of transconjugants of STEC 6-20 selected from the oxytetracycline agar plates (average transfer frequency, 10−6) were greater than the numbers of transconjugants of STEC 6-20 recovered from the tetracycline agar plates (average transfer frequency, 10−8) (Table 1). In bovine feces kept at the same temperature, however, similar numbers of transconjugants were recovered from the two types of selective agar plates for each donor strain (Table 1).
In general, the populations of STEC 6-20 transconjugants in bovine feces and storm water increased over time during the sampling period; the only exception was the transconjugants recovered from storm water and grown on tetracycline agar plates, which were detected only at day 7 (Table 1). The populations of STEC 7-63 transconjugants decreased over time during the sampling period (Table 1).
Influence of pH.
Because transconjugants were not recovered from the conjugation mixtures held at lower temperatures in the experiments described above, the influence of pH on conjugation efficiency was determined only at 28°C.
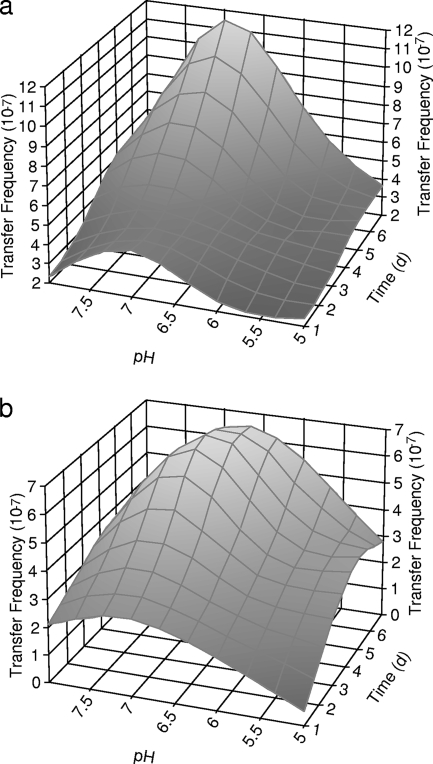
Integron-mediated antibiotic resistance genes in STEC 6-20 were transferred at higher frequencies in pH 7 storm water (Fig. 1a and 1b). The conjugation efficiencies decreased as the pH of the storm water increased or decreased from this value. Lower conjugation efficiencies for STEC 6-20 were observed at pH 5 (Fig. 1a and 1b). No transconjugants were obtained in the conjugation experiment performed with STEC 7-63 in storm water.
FIG. 1.
Frequencies of transfer of antibiotic resistance genes from STEC 6-20 to MG1655 in storm water with pH values of 5, 6, 7, and 8 at 28°C. (a) Transconjugants selected on SMAC supplemented with oxytetracycline. (b) Transconjugants selected on SMAC supplemented with tetracycline.
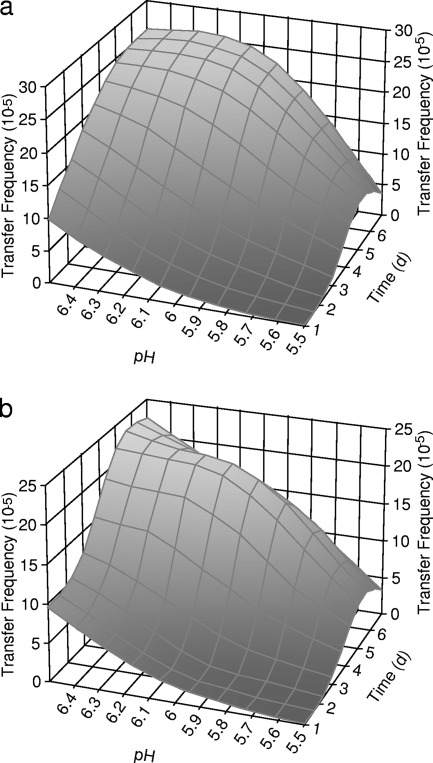
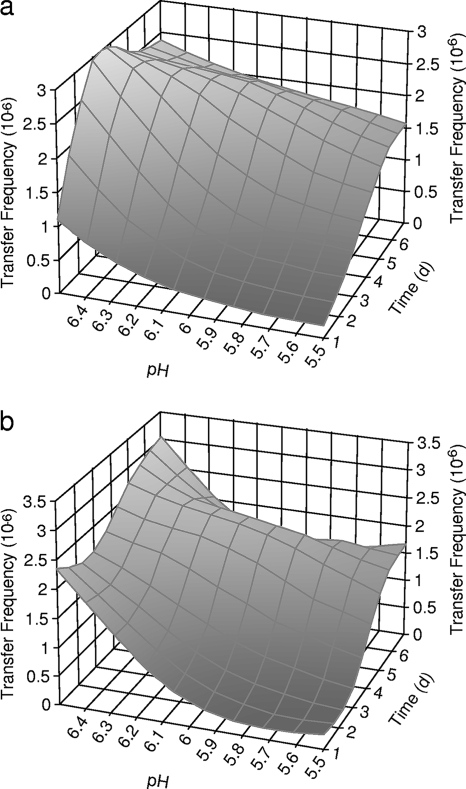
In bovine feces, integron-mediated antibiotic resistance genes were transferred from both of the STEC donors to the recipients. Similar numbers of antibiotic-resistant transconjugants were recovered from the selective agar plates supplemented with oxytetracycline and the selective agar plates supplemented with tetracycline (Fig. 2a, 2b, 3a, and 3b). Higher rates of antibiotic resistance gene transfer were found in bovine feces with a pH of 6.5 (Fig. 2 and 3). The conjugation efficiencies were lower at the lower fecal pH. The antibiotic resistance genes were transferred from both of the donor strains at lower efficiencies in bovine feces with a pH of 5.5.
FIG. 2.
Frequencies of transfer of antibiotic resistance genes from STEC 6-20 to MG1655 in bovine feces with pH values of 5.5, 6.0, and 6.5 at 28°C. (a) Transconjugants selected on SMAC supplemented with oxytetracycline. (b) Transconjugants selected on SMAC supplemented with tetracycline.
FIG. 3.
Frequencies of transfer of antibiotic resistance genes from STEC 7-63 to MG1655 in bovine feces with pH values of 5.5, 6.0, and 6.5 at 28°C. (a) Transconjugants selected on SMAC supplemented with oxytetracycline. (b) Transconjugants selected on SMAC supplemented with tetracycline.
Overall, the numbers of transconjugants derived from both of the STEC donors increased over time before day 5 and dropped slightly after this sampling time (Fig. 1, 2, and 3).
Analysis of transconjugants.
The antibiotic susceptibility test showed that the transconjugants obtained were resistant to streptomycin and sulfisoxazole, indicating that there was transfer of class 1 integron-mediated antibiotic resistance genes. The transconjugants were also resistant to tetracycline and oxytetracycline, suggesting that the genes coding for resistance to tetracycline and oxytetracycline were cotransferred with the integron-mediated antibiotic resistance genes.
DISCUSSION
Conjugation in storm water and bovine feces.
The efficiency of transfer of integron-mediated antibiotic resistance genes from both of the STEC donors was higher when conjugation took place in bovine feces. Bovine feces is semisolid and contains waste materials from animal digestive tracts, which provide support for bacterial cell contact during the conjugation process, a critical step for the transfer of genes associated with plasmids. Storm water contains much less solid waste and therefore lacks many of the conditions that favor bacterial conjugation.
Bovine feces contain larger amounts of organic materials than storm water and may therefore have more nutrients that can be utilized by bacterial cells. Previous research shown that the presence of nutrients in a conjugation mixture resulted in greater efficiency of gene transfer. Gotz and Smalla reported that the transfer rates of an IncQ plasmid were more than 10-fold greater when conjugation took place in soil supplemented with pig manure than when conjugation took place in non-manure-supplemented soil (6). It has been shown that F pilus expression is essential for formation of the mating bridge between gram-negative bacterial cells at the early stage of conjugation (4). Curtiss et al. reported that the expression of F pili was influenced by a growth medium (4). The cells of E. coli expressed a greater number of F pili in minimal growth medium supplemented with 0.5% Casamino Acids and glucose (4). However, it is worth noting that an increase in the number of transconjugants may be partially due to the growth of transconjugant cells after the transfer of antibiotic resistance genes has occurred, which may not necessarily be the result of increased conjugation efficiency.
Influence of temperature.
According to the results of the present study, incubation temperatures of 4 and 15°C did not favor the transfer of integron-mediated antibiotic resistance genes in bovine feces and storm water. Previous studies showed that a temperature higher than 15°C was required for conjugal transfer of R-plasmids between E. coli cells (21, 23). Singleton and Anson (21) found that transfer of an R-plasmid between E. coli cells occurred at 17 to 37°C when they studied the transfer of the plasmid at six different temperatures, 15, 17, 20, 22, 27, and 37°C; no transconjugants were obtained when the conjugation experiment was conducted at 15°C. In the present study, transfer of integron-mediated antibiotic resistance genes from both of the donor strains took place at 28°C, the average summer temperature in Griffin, GA. The influence of temperature on the efficiency of conjugal transfer may be related to the expression of F pili at various temperatures (18). The expression of F pili in E. coli has been shown to be regulated by temperature (18). Novotny and Lavin reported that optimal expression of F pili occurred at temperatures between 37 and 42°C, decreased at temperatures below 37°C, and diminished at temperatures below 25°C (18).
Influence of pH.
Transfer of antibiotic resistance genes occurred in bovine feces and storm water with a near-neutral pH, except for the conjugation performed with STEC7-63 in storm water. In previous studies workers observed optimum conjugation efficiencies in nutrient broth and in soil samples with a pH near 7 (22). Krasovsky and Stotzky (12) found that although the donor cells were able to survive at a pH as low as 5.7 in soil samples, conjugation occurred only when the pH was near 7. A suboptimal pH inhibited the growth of bacterial cells and lowered the frequency of DNA transfer (12). DNA could undergo DNA depurination under acidic conditions. DNA replication in recipient cells after conjugation could be suppressed by a suboptimal pH. Like what happened at 28°C, the higher numbers of antibiotic-resistant transconjugants in samples with a neutral pH may partially be influenced by the higher growth rate of the transconjugant cells.
Antibiotic-resistant STEC in an agricultural environment.
Cattle are known to be the primary reservoir for both O157 and non-O157 STEC (2). The occurrence of STEC in an agricultural environment can lead to contamination of both water and food. STEC infections have been linked to contaminated water supplies (10; http://www.fda.gov/bbs/topics/NEWS/2007/NEW01593.html). These organisms have been shown to persist for as long as 4 to 8 weeks at 25°C in bovine feces (5) and for up to 90 days in river water (25). Studies have shown that fecal excretion of E. coli O157:H7 in cattle occurred at the highest rates during the spring and late-summer months (3, 16). This time when there is a high rate of excretion generally reflects the start of a seasonal peak (spring and fall) of E. coli O157:H7 infections (11).
Transfer of antibiotic resistance genes among bacteria is a threat to both human and veterinary medicine. In this study, the transfer of integron-mediated antibiotic resistance genes in STEC isolates was demonstrated in bovine feces and storm water with a natural pH at a temperature representing the average temperature of the summer months in the Griffin, GA, area. The conjugation efficiencies of antibiotic resistance genes of the two STEC donors were different. The precise genetic basis for this difference is currently unknown. This study provided evidence of antibiotic resistance gene transfer between a pathogenic strain and a nonpathogenic strain of E. coli in simulated agricultural environments. However, it is important to note that in the present study the conjugation experiments were conducted with sterile fecal materials and storm water. The autoclaving treatment simplified the data-gathering process. The conjugation efficiencies of the antibiotic resistance genes in sterile samples could be somewhat different from those in natural, nonautoclaved samples.
Footnotes
Published ahead of print on 13 June 2008.
REFERENCES
- 1.Bell, R. G. 1997. Distribution and sources of microbial contamination on beef carcasses. J. Appl. Microbiol. 82:292-300. [DOI] [PubMed] [Google Scholar]
- 2.Bettelheim, K. A. 2000. Role of non-O157 VTEC. Symp. Ser. Soc. Appl. Microbiol. 38S-50S. [DOI] [PubMed]
- 3.Chapman, P. A., C. A. Siddons, A. T. Gerdan Malo, and M. A. Harkin. 1997. A 1-year study of Escherichia coli O157 in cattle, sheep, pigs and poultry. Epidemiol. Infect. 119:245-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Curtiss, R., L. G. Caro, D. P. Allison, and D. R. Stallions. 1969. Early stages of conjugation in Escherichia coli. J. Bacteriol. 100:1091-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fukushima, H., K. Hoshina, and M. Gomyoda. 1999. Long-term survival of Shiga toxin-producing Escherichia coli O26, O111, and O157 in bovine feces. Appl. Environ. Microbiol. 65:5177-5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gotz, A., and K. Smalla. 1997. Manure enhances plasmid mobilization and survival of Pseudomonas putida introduced into field soil. Appl. Environ. Microbiol. 63:1980-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall, R. M., and H. W. Stokes. 1993. Integrons: novel DNA elements which capture genes by site-specific recombination. Genetica 90:115-132. [DOI] [PubMed] [Google Scholar]
- 8.Hancock, D. D., T. E. Besser, D. H. Rice, E. D. Ebel, D. E. Herriott, and L. V. Carpenter. 1998. Multiple sources of Escherichia coli O157 in feedlots and dairy farms in the northwestern USA. Prev. Vet. Med. 35:11-19. [DOI] [PubMed] [Google Scholar]
- 9.Harmison, B., M. L. Eastridge, and J. L. Firkins. 1997. Effect of percentage of dietary forage neutral detergent fiber and source of starch on performance of lactating Jersey cows. J. Dairy Sci. 80:905-911. [DOI] [PubMed] [Google Scholar]
- 10.Jackson, S. G., R. B. Goodbrand, R. P. Johnson, V. G. Odorico, D. Alves, K. Rahn, J. B. Wilson, M. K. Welch, and R. Khakhria. 1998. Escherichia coli O157:H7 diarrhea associated with well water and infected cattle on an Ontario farm. Epidemiol. Infect. 120:17-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones, D. L. 1999. Potential health risks associated with the persistence of Escherichia coli O157 in agricultural environments. Soil Use Manag. 15:76-83. [Google Scholar]
- 12.Krasovsky, V., and G. Stotzky. 1987. Conjugation and genetic recombination in Escherichia coli in sterile and nonsterile soil. Soil Biol. Biochem. 19:631-638. [Google Scholar]
- 13.Mazel, D., and J. Davies. 1999. Antibiotic resistance in microbes. Cell. Mol. Life Sci. 56:742-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McEwen, S. A., and P. J. Fedorka-Cray. 2002. Antimicrobial use and resistance in animals. Clin. Infect. Dis. 34(Suppl. 3):S93-S106. [DOI] [PubMed] [Google Scholar]
- 15.McManus, P. S., V. O. Stockwell, G. W. Sundin, and A. L. Jones. 2002. Antibiotic use in plant agriculture. Annu. Rev. Phytopathol. 40:443-465. [DOI] [PubMed] [Google Scholar]
- 16.Mechie, S. C., P. A. Chapman, and C. A. Siddons. 1997. A fifteen month study of Escherichia coli O157:H7 in a dairy herd. Epidemiol. Infect. 118:17-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.NCCLS. 2003. Performance standards for antimicrobial disc and dilution susceptibility tests for bacteria isolated from animals, p. 1-64. Approved standard M31-A2, 2nd ed. NCCLS, Wayne, PA.
- 18.Novotny, C. P., and K. Lavin. 1971. Some effects of temperature on the growth of F pili. J. Bacteriol. 107:671-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pallecchi, L., C. Lucchetti, A. Bartoloni, F. Bartalesi, A. Mantella, H. Gamboa, A. Carattoli, F. Paradisi, and G. M. Rossolini. 2007. Population structure and resistance genes in antibiotic-resistant bacteria from a remote community with minimal antibiotic exposure. Antimicrob. Agents Chemother. 51:1179-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prescott, J. F. 2006. History of antimicrobial usage in agriculture: an overview, p. 19-28. In F. M. Aarestrup (ed.), Antimicrobial resistance in bacteria of animal origin. ASM Press, Washington, DC.
- 21.Singleton, P., and A. E. Anson. 1981. Conjugal transfer of R-plasmid R1drd-19 in Escherichia coli below 22°C. Appl. Environ Microbiol. 42:789-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singleton, P., and A. E. Anson. 1983. Effect of pH on conjugal transfer at low temperatures. Appl. Environ. Microbiol. 46:291-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trevors, J. T., and K. M. Oddie. 1986. R-plasmid transfer in soil and water. Can. J. Microbiol. 32:610-613. [DOI] [PubMed] [Google Scholar]
- 24.Walsh, C., G. Duffy, R. O'Mahony, S. Fanning, I. S. Blair, and D. A. McDowell. 2006. Antimicrobial resistance in Irish isolates of verocytotoxigenic Escherichia coli—VTEC. Int. J. Food Microbiol. 109:173-178. [DOI] [PubMed] [Google Scholar]
- 25.Wang, G., and M. P. Doyle. 1998. Survival of enterohemorrhagic Escherichia coli O157:H7 in water. J. Food Prot. 61:662-667. [DOI] [PubMed] [Google Scholar]
- 26.Willshaw, G. A., T. Cheasty, H. R. Smith, S. J. O'Brien, and G. K. Adak. 2001. Verocytotoxin-producing Escherichia coli (VTEC) O157 and other VTEC from human infections in England and Wales: 1995-1998. J. Med. Microbiol. 50:135-142. [DOI] [PubMed] [Google Scholar]
- 27.Yan, N. D., and R. Strus. 1980. Crustacean zooplankton communities of acidic, metal contaminated lakes near Sudbury, Ontario. Can. J. Fish. Aquat. Sci. 37:2282-2293. [Google Scholar]
- 28.Zhao, S., D. G. White, B. Ge, S. Ayers, S. Friedman, L. English, D. Wagner, S. Gaines, and J. Meng. 2001. Identification and characterization of integron-mediated antibiotic resistance among Shiga toxin-producing Escherichia coli isolates. Appl. Environ. Microbiol. 67:1558-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]