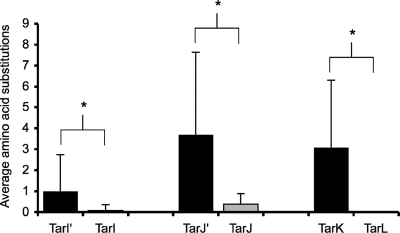
FIG. 4.
Analysis of genetic drift for genes linked to ribitol phosphate polymer formation. The translated sequences of tarI′, tarI, tarJ′, tarJ, tarK, and tarL were analyzed for congruency among S. aureus strains. Protein sequences from 13 S. aureus strains were aligned pairwise, using BLAST (National Center for Biotechnology Information [http://www.ncbi.nlm.nih.gov]), to strain COL in order to assess primary sequence variance. Alignments were performed over the entire sequence length (TarI′, 238 amino acids [aa]; TarI, 238 aa; TarJ′, 341 aa; TarJ, 341 aa; TarK, 564 aa; and TarL, 562 aa). S. aureus strains COL, Newman, Mu50, N315, JH9, JH1, Mu3, USA300_TCH1516, USA 300, NCTC 8325, RF122, MRSA252, MW2, and MSSA476 were used in this study. The data plotted show the average numbers of amino acid substitutions observed for the translated products of the loci. From these data, it can be seen that the products of the tarI′J′K gene cluster have a higher level of divergence than the products of the tarIJL gene cluster. *, P < 0.05. For the products of the tarI′J′K genes, the various numbers of substitutions as well as differing types and locations of substitutions per strain are reflected in the large standard deviation, supporting the notion that these substitutions occurred randomly. The few small sequence variations observed for products of the tarIJL gene cluster, however, were all conservative sequence substitutions common to many strains in a group. These data suggest that only the tarIJL gene cluster is stabilized by purifying selection.