Abstract
The conserved rodA and ftsW genes encode polytopic membrane proteins that are essential for bacterial cell elongation and division, respectively, and each gene is invariably linked with a cognate class B high-molecular-weight penicillin-binding protein (HMW PBP) gene. Filamentous differentiating Streptomyces coelicolor possesses four such gene pairs. Whereas rodA, although not its cognate HMW PBP gene, is essential in these bacteria, mutation of SCO5302 or SCO2607 (sfr) caused no gross changes to growth and septation. In contrast, disruption of either ftsW or the cognate ftsI gene blocked the formation of sporulation septa in aerial hyphae. The inability of spiral polymers of FtsZ to reorganize into rings in aerial hyphae of these mutants indicates an early pivotal role of an FtsW-FtsI complex in cell division. Concerted assembly of the complete divisome was unnecessary for Z-ring stabilization in aerial hyphae as ftsQ mutants were found to be blocked at a later stage in cell division, during septum closure. Complete cross wall formation occurred in vegetative hyphae in all three fts mutants, indicating that the typical bacterial divisome functions specifically during nonessential sporulation septation, providing a unique opportunity to interrogate the function and dependencies of individual components of the divisome in vivo.
Gram-positive Streptomyces spp. are actinomycetes that typically inhabit terrestrial soils and marine sediments as free-living saprophytes. In a manner similar to that of a filamentous fungus, a streptomycete colonizes its particulate environment by growing branching multigenomic hyphae that form a ramifying network, enabling the organism to exploit a localized nutrient source. Being nonmotile, they achieve dispersal by means of unigenomic spores borne on specialized nonfeeding aerial hyphae. The latter grow out of the semiaqueous environment inhabited by the feeding vegetative hyphae of terrestrial streptomycetes and ultimately undergo multiple coordinated cell divisions, generating chains of spores. The spores can then be dispersed by physical agents or the activities of motile animals inhabiting the same niche.
Streptomyces spp. grow by hyphal tip extension and subapical branching. Most de novo cell wall synthesis is at the tips, rather than by insertion of new murein into the lateral walls (19). Cross wall formation in the feeding substrate hyphae is relatively infrequent and a subapical cell separated from the tip compartment by such a cross wall can only grow once it has created a new tip by lateral branching. Tip growth contrasts markedly to the paradigm in rod-shaped bacteria such as Escherichia coli and Bacillus subtilis, which grow by intercalation of new murein into lateral walls, not into the cell wall surrounding the poles: the cell wall material of polar caps is inert after its initial synthesis during cell division (10). The mechanism for apical extension of Streptomyces hyphae is unclear. One important component of an apical protein complex has been identified: DivIVA. This protein is essential for growth, and its overexpression in preformed hyphae induces multiple branchlike lateral outgrowths, consistent with a role in stimulating tip growth (18). Interestingly, rod-shaped actinomycetes such as corynebacteria and mycobacteria grow by apical extension at both poles, suggesting that apical growth may be conserved among members of the taxon (41, 44, 50). Again, DivIVA localizes to the poles (and septa) in these unicellular bacteria and is implicated in the control of apical growth.
In a nonactinomycete gram-positive bacterium such as B. subtilis, DivIVA localizes to the poles but has an entirely different function in that it sequesters two proteins, MinC and MinD, which function as cell division inhibitors (36). Largely due to the localization of these proteins to the pole regions, vegetative septation is confined to the midcell in Bacillus spp. These cell division inhibitors and their mode of action appear to be ubiquitous among bacteria, with a notable exception being the actinomycetes that lack a bona fide Min system (19). Other aspects of cell division are remarkable in streptomycetes. One example concerns the tubulin homolog FtsZ, which polymerizes into the cytokinetic Z ring. A single ftsZ gene is required for two types of septation in Streptomyces coelicolor: infrequent cross wall formation that separates nondetached syncytial cells in the vegetative mycelium and the multiple synchronous septation of the apical compartments of aerial hyphae that leads to the formation of unigenomic spores that ultimately separate from each other (38). These developmentally distinct types of cell division are in part a consequence of how the ftsZ gene is regulated: there are at least three promoters, one of which is specifically activated in the aerial hyphae (20). In addition, the isolation of a missense mutation in ftsZ that preferentially affects sporulation septation indicates that there are differences in the mechanism of septation between vegetative and sporogenic hyphae (27). The synchronous formation of regularly spaced septa in the latter is particularly intriguing: not only is there an absence of the Min system to guide Z-ring formation but also nucleoid occlusion, which affects the formation of Z rings in E. coli and vegetative cells of B. subtilis, appears to have little influence in Streptomyces (19). The streptomyete-specific SsgA protein may have a role in marking positions in cell walls of aerial hyphae for subsequent septation to occur (42). As an early event in sporulation septation, FtsZ first polymerizes into spirals along the length of the syncytial filament; these spirals are subsequently reorganized into regularly spaced rings at the positions of subsequent septation (26). The identity of proteins that stabilize and anchor Z rings to the membrane at these positions is unclear: there are no apparent orthologs of the ZipA and FtsA proteins that perform these functions at the midcell of E. coli or ZapA and SpoIIE that have a membrane-anchoring function at the midcell and pole, respectively, during cell division and sporulation of B. subtilis (23). The functions and interactions between other components of the “divisome” in Streptomyces remain to be investigated.
In contrast to other bacteria, cell division in Streptomyces is not essential for viability. This was first demonstrated with the isolation of an ftsZ-null mutant that lacks cross walls and grows as one large syncytium (38). Here, we analyze the function of proteins that belong to the SEDS (for shape, elongation, division, and sporulation) family. These large proteins have multiple membrane-spanning domains and are believed to facilitate peptidoglycan remodeling essential for growth and cell division in other bacteria (2, 39). Genes encoding SEDS proteins are found invariably in the proximity of genes encoding class B high-molecular-weight penicillin-binding proteins (HMW PBPs), suggesting a functional relationship with these PBPs. E. coli possesses two such gene pairs, one encoding FtsW and FtsI, required for cell division, and another encoding RodA and PBP2, necessary for cell elongation. Streptomyces spp. possess four equivalent gene pairs, and our investigations reveal that one of these pairs, annotated as ftsW and ftsI, is required specifically for cell division in sporogenic aerial hyphae. The inability of FtsZ spirals to reorganize into Z rings in the aerial hyphae of ftsW or ftsI mutants indicates a pivotal early role of the FtsW protein in the assembly of divisome proteins during sporulation septation. We also reexamine the role of ftsQ, located in a gene cluster together with ftsW. Detailed cytological analysis of an ftsQ mutant suggests that the FtsQ protein is required for coordinating septal peptidoglycan synthesis with constriction of the Z ring in sporogenic hyphae. Consequently, analysis of sporulation septation in Streptomyces provides a unique insight into the in vivo function of components of the bacterial divisome.
MATERIALS AND METHODS
Bacterial strains and media.
S. coelicolor A3(2) and E. coli strains used in the present study are listed in Table 1. E. coli strain JM109 was used for cloning, and strain ET12567/pUZ8002 was used to drive the conjugative transfer of nonmethylated DNA from E. coli to S. coelicolor, as described previously (31). Cultivation of E. coli strains was performed by using standard procedures (47). S. coelicolor strains were grown on mannitol soy flour agar plates (MS agar), minimal medium, or 2×YT agar plates (31). Mutagenesis cosmids are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Transposon insertiona (position in genome) | Source or reference |
|---|---|---|---|
| Strains | |||
| S. coelicolor | |||
| M145 | Prototrophic; SCP1− SCP2− | 31 | |
| DSCO2085-1 | M145 ftsW::Tn5062(1) | SC4A10.2.H05 (2239002) | This study |
| DSCO2085-2 | M145 ftsW::Tn5062(2) | SC4A10.1.H10 (2238747) | This study |
| DSCO2085-3 | M145 ftsW::Tn5062(3) | SC4A10.2.F03 (2238425) | This study |
| DSCO2085-4 | M145 ftsW::Tn5062(4) | SC4A10.2.F08 (2238210) | This study |
| DSCO2085-5 | M145 ftsW::Tn5062(5) | SC4A10.1.F05 (2237893) | This study |
| DSCO2083 | M145 ftsQ::Tn5062 | SC4A10.2.B05 (2236542) | This study |
| DSCO2090 | M145 ftsI::Tn5062 | SC4A10.1.C09 (2246734) | This study |
| DSCO2607 | M145 sfr::Tn5062 | SCC88.2.H10 (2830098) | This study |
| DSCO3847 | M145 SCO3847::Tn5062 | SCH69.1.B04 (4232371) | This study |
| DSCO5302 | M145 SCO5302::Tn5062 | SC6G9.1.D07 (5775697) | This study |
| DSCO3846::pSHRC1 | M145::pSHRC1 rodA::Tn5062 | SCH69.1.E11 (4230660) | This study |
| DSCO5302/2607 | M145 SCO5302::Tn5062 SCO2607::Tn5062-hyg | SC6G9.1.D07 (5775697), SCC88.2.H10 (2830098) | This study |
| DSCO2085/2607 | M145 ftsW::Tn5062 SCO2607::Tn5062-hyg | SC4A10.2.H05 (2239002), SCC88.2.H10 (2830098) | This study |
| DSCO2085/5302 | M145 ftsW::Tn5062-hyg SCO5302::Tn5062 | SC4A10.2.H05 (2239002), SC6G9.1.D07 (5775697) | This study |
| E. coli | |||
| JM109 | F′ traD36 proA+B+ lacIq Δ(lacZ)M15 Δ(lac-proAB) glnV44 e14 gyrA96 recA1 relA1 endA1 thi hsdR17 | 52 | |
| ET12567(pUZ8002) | dam13::Tn9 dcm-6 hsdM hsdR recF143 zjj-201::Tn10 galK2 galT22 ara14 lacY1 xyl-5 leuB6 thi-1 tonA31 rpsL136 hisG4 tsx-78 mtlI glnV44, containing the nontransmissible oriT mobilizing plasmid pUZ8002 | 21 | |
| Plasmids | |||
| pBluescript II SK(+) | Cloning vector, ampicillin resistance | 1 | |
| pKF41 | ftsZ::EGFP | 26 | |
| pSH152 | E. coli-S. coelicolor shuttle vector, hygromycin resistance | This study | |
| pSHBW1 | pSH152 containing ftsW, murG, and ftsQ | This study | |
| pSHBW4 | pSH152 containing the ftsW gene and part of murD | This study | |
| pSHBW7 | pSH152 with ftsQ | This study | |
| pSHBW12 | pSHBW1 with in-frame deletion in ftsW | This study | |
| pSHRC1 | pSH152 containing SCO3843 to SCO3847 | This study | |
| pSFTI1 | pSH152 containing SCO2092, ftsL and ftsI | This study | |
| pSFTI3 | pSH152 containing SCO2093, SCO2092, ftsL, and ftsI | This study | |
| pUWL219 | E. coli-S. coelicolor shuttle vector, ampicillin and thiostrepton resistance | 51 |
Details of each transposon insertion can be found at http://strepdb.streptomyces.org.uk/.
Plasmid construction.
The plasmids used are listed in Table 1. DNA manipulation and cloning were carried out according to standard protocols (47). All DNA manipulations were carried out with E. coli JM109 as the host. Plasmid constructs were verified by restriction digestion and DNA sequencing. A derivative of pSET152 (4), pSH152, conferring hygromycin resistance, was engineered for genetic complementation analysis: the apramycin resistance gene (excised using the SacI-isoschizomer EcoICRI) was replaced by a blunt-ended BglII/BamHI fragment from pIJ963 (31) containing the Streptomyces hygroscopicus hygromycin resistance gene. The plasmid inserts used to complement ftsW and ftsQ mutations are depicted in Fig. S3 in the supplemental material. To construct plasmid pBSW1, a 7,478-bp EcoRI fragment from cosmid SC4A10 (46) with a Tn5062 insertion (5) at position 15834, containing ftsW, murG, and ftsQ genes, was cloned into pBluescript II SK(+) (Stratagene). From pBSW1, a 3,963-bp NotI/HindIII fragment was excised and blunt ended. This fragment was cloned into pSH152 at its unique EcoRV site to obtain plasmid pSHBW1. Plasmid pSHBW1 was digested with KpnI and AgeI and self-ligated after blunt ending to generate pSHBW12 plasmid with an in-frame deletion in the ftsW gene. Plasmid pBSW1 was digested with BamHI and self-ligated to remove the murG and ftsQ genes, leaving the ftsW gene with its putative promoter region, generating pBSW12. From pBSW12, an EcoRV/XbaI fragment was isolated and ligated with pSH152 cut with EcoRV and XbaI to obtain the ftsW complementing plasmid pSHBW4. To generate the ftsQ complementing plasmid pSHBW7, pSHBW1 was digested with BamHI, and a 4,673-bp fragment, containing ftsQ, part of murG, the origin of plasmid replication and the hygromycin resistance marker, and a 3,681-bp fragment, containing oriT and the phiC31 attachment site, were purified and religated. To engineer a strain with a second copy of rodA integrated at the phiC31 att site, a 10,362-bp SphI fragment was excised from cosmid SCH69 with a Tn5062 insertion at position 16172 (see Fig. S3 in the supplemental material). This fragment containing the genes SCO3843 to SCO3847, inclusive, and the apramycin resistance gene of Tn5062 was used as the source of a 7,036-bp XbaI fragment containing the SCO3843-to-SCO3847 region that was cloned into pSH152 to create pSHRC1. The plasmid inserts used to test complementation of the ftsI mutation are depicted in Fig. S3 in the supplemental material. A 3,953-bp EcoRI fragment containing the SCO2092, fstL, and ftsI genes was excised from cosmid SC4A10 with a Tn5062 insertion at position 24916 and cloned into pBluescript II SK(+), creating pBSI1. This region was subsequently subcloned as a 3,987-bp EcoRV-XbaI fragment into similarly digested pSH152, to create pSFTI1. The complementing DNA also containing SCO2093 was initially cloned into pSH152 as a 7,081-bp XmnI fragment that also contained the apramycin resistance gene of Tn5062, obtained from the same cosmid as described above. The Tn5062 sequence was deleted by digestion with XbaI and relegation to create pSFTI3.
Microscopy.
For light and fluorescence microscopy of vegetative hyphae, cultures were grown in the acute angle of a sterile coverslip inserted obliquely in 2×YT agar medium that supports little or no development of aerial hyphae. BODIPY FL vancomycin was used to stain nascent peptidoglycan as previously described (10). Staining of aerial hyphae was carried out by taking impressions of the cultures grown on MS agar plates. The aerial hyphae attached to coverslips were fixed and stained with fluorescein-conjugated wheat germ agglutinin (Fluo-WGA; SlowFade Light Antifade kit; Molecular Probes) for cell walls and with propidium iodide (PI; Sigma) for DNA as described previously (49). For visualization of the FtsZ-enhanced green fluorescent protein (EGFP) fusion protein in aerial hyphae, impression preparations were mounted in 50% glycerol in phosphate-buffered saline (26). For atomic force microscopy, images were obtained on air by using the tapping mode in a Dimension 3100 instrument (Nanoscope IV controller; Thermomicroscopes), as described elsewhere (13). Briefly, the strains under study were grown on MS agar plates, and glass coverslip impressions were taken at desired time intervals and subjected to imaging with standard tapping mode tips (Olympus, cantilever nominal spring constant, k = 0.064 N m−1). A scan rate of 1 Hz was used with resolutions of 512 by 512 or 1,024 by 1,024 lines. Height and phase images were simultaneously recorded. Height images provide quantitative information on sample surface topography, whereas phase images, although they do not represent the true topography of the sample, reveal a higher sensitivity to small surface features, resulting in images with greater detail. Phase images are derived from the phase angle between the actual vibration and the applied signal used to oscillate the AFM cantilever so that the tip intermittently taps the surface and reduces the lateral force that is applied during imaging. Transmission electron microscopy was performed as described previously (40).
RESULTS
Streptomyces spp. possess four genes encoding SEDS proteins, each linked with a PBP gene.
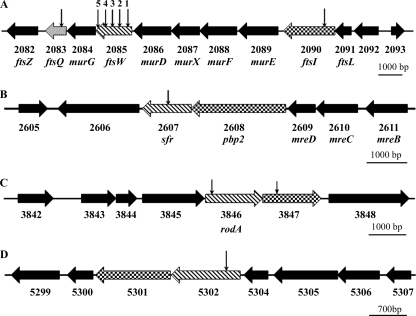
Scrutiny of the four available streptomycete genome sequences revealed four ftsW (rodA)-type genes in all species, each SEDS gene being closely linked with a gene encoding a class B HMW PBP. This is exemplified by S. coelicolor (Fig. 1). Two of these gene pairs are conserved in other actinomycetes, including rod-shaped mycobacteria, corynebacterium species, and the filamentous sporulating Saccharopolyspora erythraea, which possess no other ftsW (rodA) homologs. Consistent with the annotation of the respective mycobacterial genes, we subsequently refer to SCO3846 as rodA and SCO2085 as ftsW. The gene contexts of rodA and ftsW are highly conserved in actinomycetes, with the former being close to oriC and linked to crgA, implicated in coordinating growth and cell division in Streptomyces (13, 14). The ftsW gene is part of the well-conserved prokaryotic division cell wall (DCW) gene cluster that includes ftsZ. Interestingly, alignment with B. subtilis SEDS proteins revealed that FtsW from S. coelicolor (FtsWSc) shows the greatest homology with the sporulation-specific SEDS protein SpoVE (31% identity, 50% similarity); in the B. subtilis genome, spoVE is part of the DCW cluster, whereas ftsW, involved in vegetative cell division, is located elsewhere. Hydropathy plots (32) of the S. coelicolor and Mycobacterium tuberculosis SEDS proteins indicate that they are all polytopic membrane proteins (see Fig. S1 in the supplemental material). Both RodASc and RodAMt (RodA from M. tuberculosis) are predicted to possess 12 transmembrane (TM) domains, whereas FtsWSc has 8 and FtsWMt has 10 predicted TM domains. Topological analyses of SEDS proteins from other bacteria indicate that these proteins have in general 10 TM domains (22, 34, 45). As has previously been noted (11), FtsWMt is predicted to have a long (∼120-amino-acid) cytoplasmic C-terminal tail not present in the E. coli ortholog. The equivalent region of FtsWSc is shorter (∼50 amino acids; see Fig. S1 and S2 in the supplemental material). The two other streptomycete-specific putative SEDS proteins, SCO5302 and SCO2607, are predicted to have 12 and 10 TM domains, respectively. The SCO2607 gene (sfr) is linked with mre genes whose products are implicated in determining cell shape (9). Of the four S. coelicolor SEDS proteins, Sfr is most closely related to RodA of E. coli (30% identity, 43% similarity). SCO5302 shares considerable homology with RodASc (58% identity, 70% similarity).
FIG. 1.
Genetic organization of the four SEDS protein genes of S. coelicolor. For each locus, the SEDS protein gene is indicated by hatched shading, and the cognate PBP gene is indicated by checkered shading. SCO gene numbers and, where appropriate, gene names are shown below each diagram. The positions of each Tn5062 insertion in mutants investigated in the present study are indicated by arrows above the corresponding gene (additional details for these insertions are provided in Table 1). (A) The DCW gene cluster containing ftsW and ftsI. ftsQ is indicated by dotted shading. (B) sfr (SCO2607) gene locus adjacent to the mre genes. (C) rodA gene locus. (D) Locus containing SCO5302.
Mutagenesis of the S. coelicolor SEDS protein genes.
Transposon insertion mutants for three SEDS protein genes were constructed by using mutated cosmids generated by in vitro transposition (5). To construct these mutants, we chose Tn5062 insertions close to the start of the respective genes (Table 1; the insertion at 2239002 was chosen initially to study ftsW function) with the knowledge that there would be little likelihood of any remaining gene function in the mutants. For successful mutant construction, confirmed in representative clones by Southern hybridization, apparent allelic replacement and loss of the Supercos-1 vector (see Materials and Methods) occurred in more than 42% and up to 96% of exconjugants. In contrast, we were unable to construct a rodA mutant (the frequency of apparent allelic replacement being <1%, and with hybridization patterns in isolated clones being inconsistent with genuine allelic replacement) unless a second copy of this gene was integrated at the phiC31 att site. Mutants of the downstream class B HMW PBP gene, SCO3847, could be constructed easily in the haploid wild-type strain, although they exhibited no macroscopic phenotype. A similar observation was made for the equivalent gene of Streptomyces griseus (30). These results indicate that, whereas rodA is essential for viability, its cognate class B transpeptidase is not. Disruption of either the SCO5302 or SCO2607 gene (sfr) resulted in no discernible macroscopic phenotype with the growth conditions we tested, although spores of the SCO2607 mutant exhibited heat and detergent sensitivity (to be published). In contrast, ftsW mutants had a white (Whi) phenotype when grown on sporulation media (Fig. 2A), a finding indicative of a defect in differentiation of aerial hyphae. This Whi phenotype could be attributed to polar effects of the transposon insertion on cotranscribed downstream genes. This was investigated by genetic complementation. The developmental phenotype of the ftsW mutants was restored by inserting a complementing plasmid, pSHBW4, carrying only the ftsW gene, but was not restored by a plasmid, pSHBW12, carrying three genes, ftsW-murG-ftsQ, containing an in-frame deletion in ftsW (Fig. 2A and see Fig. S3 in the supplemental material), inferring that there are no polar effects of the ftsW transposon insertions on the downstream murG and ftsQ genes.
FIG. 2.
Phenotypic and complementation analysis of ftsW, ftsI, and ftsQ white mutants. Strains were grown on SFM sporulation medium for 3 days. The gray sporulating wild-type strain, M145, is indicated as W. Maps of the DNA fragments used for complementation analysis are provided in Fig. S3 in the supplemental material. (A) 1, DSCO2085-1 [ftsW::Tn5062(1)]; 2, DSCO2085-1 with plasmid pSHBW4; 3, DSCO2085-1 with plasmid pSHBW1; 4, DSCO2085-1 with plasmid pSHBW12; 5, DSCO2085-5 [ftsW::Tn5062(5)]. (B) 1, DSCO2090 (ftsI::Tn5062); 2, DSCO2090 with plasmid pSFTI1; 3, DSCO2090 with plasmid pSFTI3; 4, DSCO2083 (ftsQ::Tn5062); 5, DSCO2083 with plasmid pSHBW12; 6, DSCO2083 with plasmid pSHBW7.
Having confirmed that the developmental phenotype was attributable to loss of FtsW function, we subsequently constructed mutants to address whether the cognate HMW PBP, FtsI, was also involved in sporulation. All Tn5062 mutants of ftsI showed a similar Whi phenotype that could be restored by introducing a complementing plasmid, pSFTI3, containing a putative operon consisting of the upstream SCO2092, ftsL, and ftsI genes and the divergently transcribed SCO2093 gene (Fig. 2B and see Fig. S3 in the supplemental material). A shorter DNA fragment cloned into pSFTI1, lacking the SCO2093 gene and containing 449 bp of the intergenic sequence upstream of the GTG start codon of SCO2092, could not complement the ftsI mutation. This could imply that a critical promoter for the operon containing ftsI is located some distance upstream, reminiscent of the dependence of expression of E. coli ftsI on a promoter located 1.9 kb upstream of the gene itself (28).
FtsW and FtsI are required for Z-ring formation during sporulation septation in S. coelicolor.
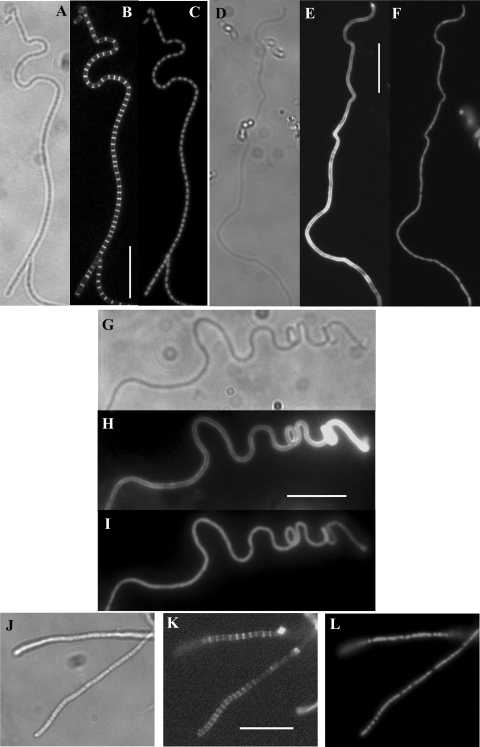
To examine the cytological effects of the various mutations, the formation of septa and segregation of DNA in aerial hyphae was examined after staining cell walls with Fluo-WGA and chromosomal DNA with PI. Sporulation septa were formed at regular intervals in the aerial hyphae of the wild type (Fig. 3 A to C) and the SCO5302 or SCO2607 (sfr) mutants, these septa defining prespore compartments each containing a single condensed nucleoid. In contrast, the aerial hyphae of both the ftsW (Fig. 3D to F) and ftsI (Fig. 3G to I) mutants remained syncytial, with no apparent chromosome condensation, even after prolonged incubation. Some of these hyphae exhibited coiling.
FIG. 3.
Analysis of sporulation septation in the wild type and ftsW, ftsI, and ftsQ mutants. Phase-contrast (A) and Fluo-WGA-stained (B) and PI-stained (C) images of aerial hyphae of strain M145 after 48 h of growth showing regular septation and condensed nucleoids in each prespore compartment. In contrast, there is an absence of septation and no chromosomal condensation in panels E and F (Fluo-WGA and PI stained, respectively) showing aerial hyphae of DSCO2085-1 [ftsW::Tn5062(1)] after 96 h of growth and in panels H and I (Fluo-WGA and PI stained, respectively) showing aerial hyphae of DSCO2090 (ftsI::Tn5062) after 96 h of growth. (K and L) Fluo-WGA-strained (K) and PI-stained (L) aerial hyphae of DSCO2083 (ftsQ::Tn5062) after 80 h of growth showing delayed and less-regular septation and only partial nucleoid condensation. Panels D, G, and J are phase-contrast images for the ftsW, ftsI, and ftsQ mutants, respectively. Scale bar, 10 μm.
To attempt to define the earliest stage in septum formation dependent on both FtsW and FtsI function, we expressed EGFP-tagged FtsZ in the mutants and compared its distribution to that in the wild type. In the latter, a typical progression was observed with aerial hyphae sampled from earlier time points containing diffuse fluorescence typical of nonassembled FtsZ-EGFP or occasionally the protein assembled into spirals, and hyphae in older samples containing either spirals or multiple regular-spaced rings (Fig. 4A and B) or having progressed through cell division. In contrast, the aerial hyphae of the ftsW (Fig. 4C and D) and ftsI (Fig. E and F) mutants examined over an extended period contained only diffuse fluorescence or FtsZ-EGFP spirals, but no rings.
FIG. 4.
Assembly of FtsZ::EGFP in aerial hyphae of the wild type and the ftsW, ftsI, and ftsQ mutants. Representative images of the distribution of FtsZ::EGFP are shown alongside the corresponding phase-contrast image. Regular FtsZ rings could be observed in aerial hyphae of strain M145 (B) grown for 40 h but not in DSCO2085-1 [ftsW::Tn5062(1); panel D] or DSCO2090 (ftsI::Tn5062; panel F) grown for extended periods, sampled here at 84 h, in which spirals or diffuse fluorescence were observed. Delayed ring formation was apparent in DSCO2083 (ftsQ::Tn5062; panel H) grown for 76 to 78 h. Scale bar, 10 μm.
These data are consistent with an FtsW-FtsI complex either directly providing a membrane anchor to stabilize FtsZ rings or having an indirect role in this process. An in vitro interaction between FtsWMt and FtsZMt is dependent on the long ∼120-amino-acid cytoplasmic C-terminal tail of the former (11). Although the equivalent region of FtsWSc is substantially shorter, we attempted to define a minimal FtsWSc necessary to allow Z-ring formation and subsequent cell division by analyzing mutants with Tn5062 insertions at different positions within the gene (Table 1 and Fig. 1). Insertions that resulted in truncated proteins lacking any of the eight predicted TM domains were similar in phenotype to the initial mutant we examined in that they were white and aerial hyphae lacked sporulation septa. When FtsZ-EGFP was expressed in these mutants, we could not detect Z-ring formation. All of these mutants were complemented by plasmid pSHBW4 containing the wild-type ftsW. In contrast, a mutant with a C-terminal insertion at position 2237893 encoding a truncated protein lacking the last 18 amino acids of the predicted cytoplasmic tail had a normal gray phenotype (Fig. 2A). The truncated FtsW protein ends with an 11-amino-acid sequence encoded by one end of Tn5062, TVSYTHLNHHR, that does not align with the native C-terminal sequence (see Fig. S2 in the supplemental material). Z-ring formation was evident in the aerial hyphae of this mutant, which differentiated normally into spore chains.
FtsQ functions during sporulation septation after stabilization of Z rings.
The pivotal role of both FtsW and FtsI in stabilizing Z rings in the aerial hyphae could imply that concerted assembly of all or part of the divisome is necessary for Z-ring formation. An obvious component to test whether this is indeed the case was FtsQ, encoded by the third and last gene of the cluster that commences with ftsW. Allelic exchange to introduce a Tn5062 insertion near the beginning of the coding sequence occurred at high frequency (77% of the excongugants), indicating no deleterious effects of the mutation on overall growth. All ftsQ mutant clones exhibited a Whi phenotype on sporulation medium (Fig. 2B) and normal antibiotic production on minimal medium. The phenotype was restored by genetic complementation with plasmid pSHBW1 carrying the complete gene cluster consisting of ftsW, murG, and ftsQ or by pSHBW12 with the ftsW in-frame deletion (Fig. 2B and see Fig. S3 in the supplemental material). A plasmid, pSHBW7, lacking ftsW and part of murG could also complement, although the strain developed dark gray spores slightly later than the wild type or mutant complemented with either pSHBW1 or pSHBW12. This could indicate that expression of ftsQ in pSHBW7 is affected due to the loss of one or more upstream promoters.
The Whi phenotype of ftsQ mutants indicated a specific role for FtsQ during sporulation septation. After expression of the FtsZ-EGFP translational fusion in the mutants, delayed and, to an extent, less-regular Z-ring assembly was observed in the mutant aerial hyphae (Fig. 4G to H), indicating that FtsQ is not essential for Z-ring assembly. Indeed, microscopy of Fluo-WGA-stained aerial hyphae from 3-day-old cultures revealed evidence for delayed septal peptidoglycan synthesis at regular-spaced intervals (Fig. 3J to L). However, PI staining of the mutant revealed many regions of the hyphae containing diffuse nucleoids that were apparently uninterrupted by septa at the positions of the Fluo-WGA-stained material, in contrast to the regular condensed uninucleoid prespore compartments of the wild type where PI staining along the length of each hypha was clearly interrupted at the position of each septum (Fig. 3J to L). In certain locations along the length of an ftsQ mutant aerial hypha, however, it was evident that non-PI-staining areas coincided with Fluo-WGA-staining material, a finding indicative of possible genuine septation in these positions.
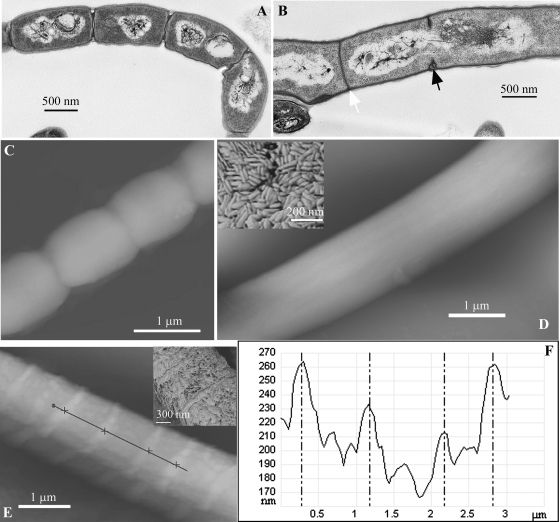
The aberrant pattern of septation was further investigated by transmission electron microscopy of aerial hyphae (Fig. 5A and B). Two types of septal structure were evident in the ftsQ mutant. There were many incomplete “thick” sporulation septa formed at regular intervals but not defining separate compartments; these partially formed septa were spanned by nucleoids. A second type was infrequent complete “thin” septa resembling those of vegetative hyphae. In contrast, the wild-type aerial hyphae possessed multiple regularly placed sporulation septa. We also examined the surface of the aerial hyphae by atomic force microscopy, visualizing the topography of the hyphal surfaces (Fig. 5C to E). In all mutants the aerial hyphae were coated with rodlet layers observed on the wild-type aerial hyphae (13). In contrast to the smooth, uninterrupted rodlet layers coating hyphae of ftsW or ftsI mutants or the localized disruption of these layers coinciding with indentations at the positions of in-growing septa that can be observed during sporulation septation in wild-type aerial hyphae (13), the ftsQ mutant possessed temporary extruded deformations or annuli, 25 to 40 nm in height, of the rodlet layers at intervals consistent with the distances between in-growing septa (Fig. 5E and F). Phase-contrast microscopy of aerial hyphae of the ftsQ mutant sampled at later time points (8 days) revealed that they exhibited lateral branches that are not formed in the wild type.
FIG. 5.
High-resolution imaging of aerial hyphae of the wild type and ftsW and ftsQ mutants. Transmission electron microscopy was used to examine sporulation septation in strain M145 after 48 h of growth, with regular thick sporulation septa and condensed nucleoids (A), and DSCO2083 (ftsQ::Tn5062) after 78 h of growth with frequent incomplete thick sporulation septa (black arrowhead), often spanned by a nucleoid, and infrequent thin cross walls (white arrowhead) (B). (C to E) Atomic force microscopy revealed differences in the external features of the hyphae: “height” images of M145 (48 h of growth), DSCO2085-1 [ftsW::Tn5062(1), 96 h of growth], and DSCO2083 (ftsQ::Tn5062, 78 h of growth) reveal a smooth morphology of the ftsW hyphae compared to the indented hyphae of septating wild-type hyphae and extruded annuli present on the surface of the ftsQ mutant. The insets in panels D and E are phase-contrast images revealing the rodlet layers of the ftsW and ftsQ mutants. (F) Graph depicting the height profile of a section along midline of the hypha, indicated by the black line in panel E.
Septation in aerial and vegetative hyphae occurs by different mechanisms.
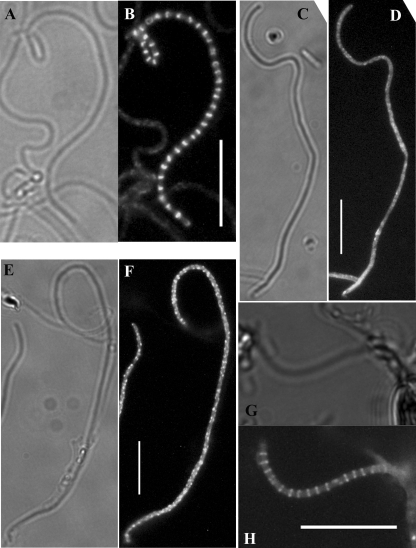
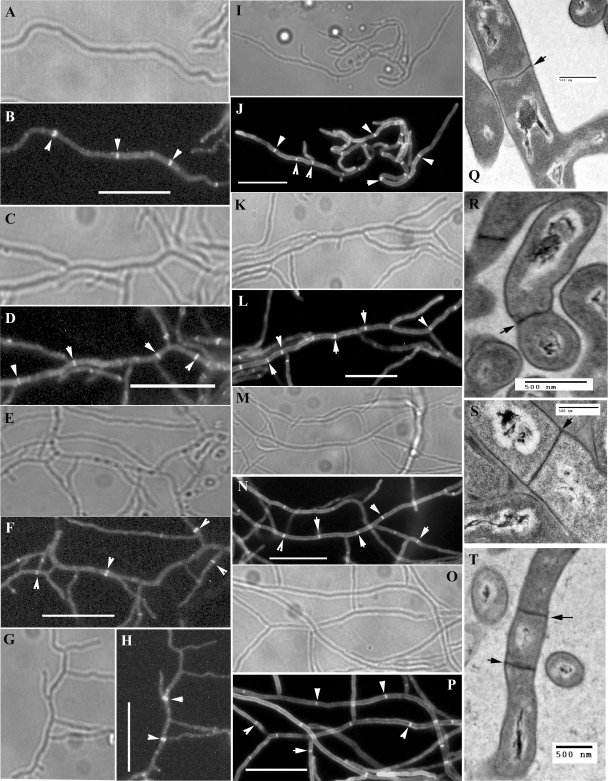
An ftsZ mutant of S. coelicolor is syncytial, lacking septa in the vegetative hyphae, and is unable to develop aerial hyphae (38). Overall, the growth of this mutant is affected, since it attains only small colony sizes, and it produces copious amounts of actinorhodin. In contrast, the ftsW, ftsI, and ftsQ mutants we constructed grew well and underwent development of aerial hyphae and with no apparent increase in antibiotic production compared to the wild type. Given these differences, we then examined the different stages of vegetative septation in these mutants. Early stages, involving Z-ring formation (Fig. 6A to H), and subsequent de novo cell wall synthesis (Fig. 6I to P) were similar in ftsW, ftsI, and ftsQ mutants and the wild type. Moreover, transmission electron microscopy of vegetative hyphae revealed complete cross walls in these mutants (Fig. 6Q to T). This indicated that the functions of the three cell division genes belonging to the DCW cluster are absolutely required for sporulation septation but not during vegetative septation. Normal vegetative septation was also observed in strains with mutations in the other two nonessential SEDS genes: SCO5302 and SCO2607 (results not shown). To exclude functional redundancy, we constructed three double mutants, one with insertions in both ftsW and SCO2607, the second with disrupted copies of both SCO5302 and SCO2607, and the third with mutations in ftsW and SCO5302; we observed normal vegetative septation in all three double mutants (results not shown).
FIG. 6.
Complete vegetative cross wall formation in mutants. (A to H) Substrate hyphae from 48-h cultures of strains expressing EGFP-tagged FtsZ were imaged to reveal Z rings, indicated by white arrowheads. Phase-contrast and fluorescence microscopy images of M145 (wild type) (A and B), DSCO2085-1 [ftsW::Tn5062(1)] (C and D), DSCO2090 (ftsI::Tn5062) (E and F), and DSCO2083 (ftsQ::Tn5062) (G and H) are shown. (I to P) Substrate hyphae from 48-h cultures stained with Fluo-vancomycin to reveal septa, indicated by white arrowheads. Phase-contrast and fluorescence microscopy images of M145 (wild type) (I and J), DSCO2085-1 [ftsW::Tn5062(1)] (K and L), DSCO2090 (ftsI::Tn5062) (M and N), and DSCO2083 (ftsQ::Tn5062) (O and P) are shown. Scale bar, 10 μm for panels A to P. (Q to T) Transmission electron microscopy images of substrate hyphae to reveal complete cross walls (black arrowheads) of M145 (Q), DSCO2085-1 [ftsW::Tn5062(1)] (R), DSCO2090 (ftsI::Tn5062) (S), and DSCO2083 (ftsQ::Tn5062) (T) are shown.
DISCUSSION
Members of the SEDS family are believed to be present in all eubacteria that synthesize peptidoglycan as part of their cell envelope. The E. coli rodA and ftsW genes and the spoVE gene of B. subtilis encode membrane proteins that control peptidoglycan synthesis during cellular elongation, division, and sporulation, respectively (29). Although rodA and ftsW are essential genes in both E. coli and B. subtilis, the B. subtilis spoVE gene is dispensable for growth and is required only for the synthesis of the spore cortex peptidoglycan. In both of these rod-shaped bacteria, RodA is required for the intercalation of the new peptidoglycan into lateral walls during growth (2), whereas FtsW is vital for synthesis of new peptidoglycan needed for formation of a new cell wall that separates daughter cells at division (39). In B. subtilis, SpoVE localizes to the forespore at an early stage of engulfment, and FtsW localizes to both vegetative and sporulation septa (45). ftsW is one of a number of highly conserved and closely associated genes that together form the DCW cluster in most eubacteria and whose products are believed to constitute the core of the bacterial cell division machinery. The broad phylogenetic distribution of the cluster suggests that the cell division machinery is of ancient origin and evolved in parallel with the cell wall and its biosynthetic pathways.
A survey of actinomycete genomes indicates conservation of the DCW cluster, including ftsW, and that, as a minimum, they contain at least one additional conserved SEDS gene, rodA. The latter is part of a conserved actinomycete gene cluster located close to oriC that includes genes encoding a cognate penicillin-binding protein and CrgA, implicated in coordinating growth and cell division in streptomycete aerial hyphae (13, 14). Given that rodA is essential for growth in other eubacteria, our inability to create a null mutant in S. coelicolor suggests a similar role in actinomycetes. A critical difference is that de novo peptidoglycan synthesis in actinomycetes occurs primarily at the poles (or hyphal tips in filamentous actinomycetes), rather than the lateral walls (19). Hence, we predict that RodA may form part of an apical protein complex, along with DivIVA, that promotes growth in actinomycetes.
The process of cross wall formation in vegetative hyphae is apparently not affected in mutants of the three nonessential S. coelicolor SEDS genes or in double mutants. Disruption of ftsW function specifically affects sporulation septation. Indeed, the vegetative cross wall formation we observed in mutants of three different fts genes confirms that the two types of septation in streptomycetes are mechanistically very distinct, as originally suggested from the functions of different forms of FtsZ (27). Functional redundancy of SEDS proteins may contribute to the formation of vegetative cross walls in the mutants we have analyzed, and we are currently constructing a triple mutant to knockout function of all three nonessential genes while also analyzing subcellular localization of the proteins. Sporulation septation is a developmentally programmed type of cell division that effectively transforms a multicellular filamentous growth style into a single-celled state reminiscent of most other eubacteria. Our data imply that sporulation septation in streptomycetes is mechanistically equivalent to vegetative septation in nonfilamentous bacteria. An important difference compared to other bacteria, however, is that sporulation septation is nonessential for viability of Streptomyces and consequently a good system for investigating functional roles of the various cell division proteins in vivo.
In E. coli, FtsW and FtsI are believed to be recruited late to the divisome in a quasilinear assembly sequence involving, first, stabilization of the Z ring with FtsA and the membrane anchor ZipA and then subsequent recruitment of FtsK and a trimeric complex involving FtsL, FtsB, and FtsQ (7, 8, 23-25). Systematic bacterial two-hybrid assays support subsequent recruitment of FtsW and FtsI, which both interact with FtsQ (15). A fragment of the periplasmic domain of FtsQ, spanning residues 67 to 75, interacts with FtsW (16), whereas the TM segment contributes to localization of FtsQ to the division site (48). In contrast, the late division proteins appear to be recruited in a concerted manner in B. subtilis (17); mutation or depletion of any one prevents all of the others assembling after formation of the Z ring, and consequently, there is no peptidoglycan synthesis at the division site. Our genetic and cytological data support a pivotal role for FtsW, both in likely recruitment of the cognate PBP FtsI and in formation of the Z ring—either acting as a membrane anchor and stabilizing factor itself or recruiting one or more additional proteins that perform this function in the absence of FtsA, ZipA, or ZapA orthologs. Chromosome condensation is also affected, implying that this process is dependent on early events in divisome formation. Loss of ftsQ function results in a block at a later stage in sporulation septation. Sporulation septal peptidoglycan synthesis begins at the division sites in ftsQ mutants, but these septa fail to complete their centripetal growth. This suggests a critical role of FtsQSc in linking constriction of the cytosolic Z-ring with invagination of newly synthesized murein. In the absence of this function, new peptidoglycan synthesis at division sites may cause localized temporary outward deformations of the surface rodlet layers, as we observed using atomic force microscopy. The temporary localized breakage of the rodlet layer at the sites of in-growing septa that is a normal event during sporulation of the wild type (13) was not observed for the ftsQ mutant, suggesting that this is a late event, presumably occurring after completion of septal in-growth. Moreover, another consequence of the failure to close sporulation septa appears to be the formation of occasional vegetative septa which may trigger subsequent branch formation in the aerial hyphae. Branching of the aerial hyphae in a ftsQ mutant blocked at an undefined stage of sporulation septation was previously reported (37). In some other respects the ftsQ mutant we describe differs from the previously described mutant, which was reported to exhibit copious actinorhodin production and reduced vegetative septation. These discrepancies may be due to strain differences and the way the respective mutants were constructed. However, the authors of that study suggested that the phenotype of their mutant, created by deleting the majority of the ftsQ coding sequence, could be in part due to the result of polarity on expression of the downstream ftsZ, although the defect in sporulation septation was “substantially restored” when the mutant was complemented by the ftsQ gene (37). In this regard, it is interesting that expression of ftsZ in the related actinomycete Corynebacterium glutamicum is controlled by at least five promoters, three of which are located in the coding region of the upstream ftsQ gene (35), while in S. griseus one of four ftsZ transcripts is initiated in the coding sequence of ftsQ (33), and there is transcriptional readthrough from within or upstream of ftsQ in S. coelicolor (20). In E. coli, FtsQ is recruited to the divisome as part of a trimeric complex with FtsL and FtsB. ftsL and divIC mutants of S. coelicolor are conditionally defective in completion of septal closure in the aerial hyphae (3), with the phenotype being apparent when osmolyte was included in the growth medium. This conditionality contrasts with the absolute defect in our ftsQ mutants, implying a more critical role for FtsQ in septum closure.
Using a combination of genetic and cytological approaches, a role for FtsW in stabilization of Z rings has been reported in E. coli (6). However, subsequent biochemical analysis has not provided evidence for a direct interaction between these proteins, and FtsW is now considered a late recruit to the divisome in this bacterium, after Z-ring formation. This may highlight how different assays, one functional (albeit based on conditional gene expression) and the other based on interactions between suspected protein partners, can lead to different conclusions. Interestingly, however, binding has been demonstrated between the cytoplasmic C-terminal tail of mycobacterial FtsW and the C-terminal portion of FtsZ (11). Indeed, in merodiploids expressing both wild-type and fluorophore-tagged versions of both proteins, this interaction helps to direct colocalization of both proteins to the septum in Mycobacterium smegmatis (43). Due to the proportion of septal Z rings scoring positive for either tagged protein, it was interpreted that, just as in E. coli, FtsW is recruited late to the divisome in mycobacteria. This interpretation is inconsistent with our observations implicating an earlier pivotal role for FtsW in stabilizing Z rings in the aerial hyphae of S. coelicolor. This could imply a fundamental difference in the assembly of divisomes in these related actinomycetes or, alternatively, may be due to dissimilarity in the experimental approaches needed to study cell division in both organisms. Both ftsW and ftsZ are essential for cell viability in mycobacteria, necessitating construction of merodiploid strains to examine localization of fluorophore-tagged proteins that may not retain full biological function. Antisense depletion of FtsW in M. smegmatis leads to irregular Z-ring assembly and prevents septation (12), which is more consistent with the more pivotal role of this SEDS protein in actinomycete cell division that we report here. The interaction between the mycobacterial proteins is believed to depend on a cluster of four aspartate residues located at the C terminus of FtsZ and an oppositely charged arginine-rich region in the cytoplasmic C terminus of FtsW (11). A cluster of aspartates is not present in the C terminus of FtsZ of S. coelicolor. Moreover, the C-terminal 18-amino-acid region of FtsWSc containing several arginine residues could be replaced with no effect on Z-ring stabilization, indicating that the nature of any direct or indirect interaction between the streptomycete proteins is unlike that described for the mycobacterial proteins. In contrast, two proline residues in two extracytoplasmic loops of mycobacterial FtsW, Pro306 and Pro386, that are required for interaction with mycobacterial PBP3 (FtsI [12]) are conserved in FtsWSc (Pro302 and Pro382; see Fig. S3 in the supplemental material). It has been suggested that the interaction between mycobacterial FtsZ and FtsW may modulate the latter's interaction with PBP3 (12). Our data imply that the formation of a complex between FtsWSc and FtsISc is required to stabilize Z rings at division sites. We are currently investigating the role of FtsWSc in Z-ring stabilization in more detail.
An important implication of these studies is that the highly conserved DCW cluster in streptomycetes primarily functions during sporulation septation. Previously, an 85-kDa PBP was suggested to function specifically during sporulation, since cefoxitin, which binds this protein, inhibits sporulation septation but not vegetative septation in submerged cultures of S. griseus (33). As we have demonstrated, this nonessential developmental cell division process offers an ideal model for genetic dissection to investigate function of the individual components of the actinomycete divisome. This can reveal vital new information to complement existing biochemical functional data and also prompt investigations into unexpected functional interactions. With this knowledge, we are continuing to probe the function of several other cell division genes.
Supplementary Material
Acknowledgments
This study was supported by grants from the European Commission (LSHM-CT-2004-005224) and the BBSRC (BB/E019242/1).
We are grateful to Klas Flärdh for the gift of plasmid pKF41.
This paper is dedicated to the memory of Kate D'Lima.
Footnotes
Published ahead of print on 13 June 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alting-Mees, M. A., and J. M. Short. 1989. pBluescript II: gene mapping vectors. Nucleic Acids Res. 179494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Begg, K. J., B. G. Spratt, and W. D. Donachie. 1986. Interaction between membrane proteins PBP3 and RodA is required for normal cell shape and division in Escherichia coli. J. Bacteriol. 1671004-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett, J. A., R. M. Aimino, and J. R. McCormick. 2007. Streptomyces coelicolor genes ftsL and divIC play a role in cell division but are dispensable for colony formation. J. Bacteriol. 1898982-8992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 11643-49. [DOI] [PubMed] [Google Scholar]
- 5.Bishop, A., S. Fielding, P. Dyson, and P. Herron. 2004. Systematic insertional mutagenesis of a streptomycete genome: a link between osmoadaptation and antibiotic production. Genome Res. 14893-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle, D. S., M. M. Khattar, S. G. Addinall, J. Lutkenhaus, and W. D. Donachie. 1997. ftsW is an essential cell-division gene in Escherichia coli. Mol. Microbiol. 241263-1273. [DOI] [PubMed] [Google Scholar]
- 7.Buddelmeijer, N., and J. Beckwith. 2002. Assembly of cell division proteins at the Escherichia coli cell center. Curr. Opin. Microbiol. 5553-557. [DOI] [PubMed] [Google Scholar]
- 8.Buddelmeijer, N., and J. Beckwith. 2004. A complex of the Escherichia coli cell division proteins FtsL, FtsB, and FtsQ forms independently of its localization to the septal region. Mol. Microbiol. 521315-1327. [DOI] [PubMed] [Google Scholar]
- 9.Burger, A., K. Sichler, G. Kelemen, M. Buttner, and W. Wohlleben. 2000. Identification and characterization of the mre gene region of Streptomyces coelicolor A3(2). Mol. Gen. Genet. 2631053-1060. [DOI] [PubMed] [Google Scholar]
- 10.Daniel, R. A., and J. Errington. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113767-776. [DOI] [PubMed] [Google Scholar]
- 11.Datta, P., A. Dasgupta, S. Bhakta, and J. Basu. 2002. Interaction between FtsZ and FtsW of Mycobacterium tuberculosis. J. Biol. Chem. 27724983-24987. [DOI] [PubMed] [Google Scholar]
- 12.Datta, P., A. Dasgupta, A. K. Singh, P. Mukherjee, M. Kundu, and J. Basu. 2006. Interaction between FtsW and penicillin-binding protein 3 (PBP3) directs PBP3 to mid-cell, controls cell septation, and mediates the formation of a trimeric complex involving FtsZ, FtsW, and PBP3 in mycobacteria. Mol. Microbiol. 621655-1673. [DOI] [PubMed] [Google Scholar]
- 13.Del Sol, R., I. Armstrong, C. Wright, and P. Dyson. 2007. Characterization of changes to the cell surface during the life cycle of Streptomyces coelicolor: atomic force microscopy of living cells. J. Bacteriol. 1892219-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Del Sol, R., A. Pitman, P. Herron, and P. Dyson. 2003. The product of a developmental gene, crgA, that coordinates reproductive growth in Streptomyces belongs to a novel family of small actinomycete-specific proteins. J. Bacteriol. 1856678-6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Lallo, G., M. Fagioli, D. Barionovi, P. Ghelardini, and L. Paolozzi. 2003. Use of a two-hybrid assay to study the assembly of a complex multicomponent protein machinery: bacterial septosome differentiation. Microbiology 1493353-3359. [DOI] [PubMed] [Google Scholar]
- 16.D'Ulisse, V., M. Fagioli, P. Ghelardini, and L. Paolozzi. 2007. Three functional subdomains of the Escherichia coli FtsQ protein are involved in its interaction with the other division proteins. Microbiology 153124-138. [DOI] [PubMed] [Google Scholar]
- 17.Errington, J., R. A. Daniel, and D. J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 6752-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flardh, K. 2003. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2). Mol. Microbiol. 491523-1536. [DOI] [PubMed] [Google Scholar]
- 19.Flardh, K. 2003. Growth polarity and cell division in Streptomyces. Curr. Opin. Microbiol. 6564-571. [DOI] [PubMed] [Google Scholar]
- 20.Flardh, K., E. Leibovitz, M. J. Buttner, and K. F. Chater. 2000. Generation of a non-sporulating strain of Streptomyces coelicolor A3(2) by the manipulation of a developmentally controlled ftsZ promoter. Mol. Microbiol. 38737-749. [DOI] [PubMed] [Google Scholar]
- 21.Flett, F., V. Mersinias, and C. P. Smith. 1997. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol. Lett. 155223-229. [DOI] [PubMed] [Google Scholar]
- 22.Gerard, P., T. Vernet, and A. Zapun. 2002. Membrane topology of the Streptococcus pneumoniae FtsW division protein. J. Bacteriol. 1841925-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goehring, N. W., and J. Beckwith. 2005. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr. Biol. 15R514-R526. [DOI] [PubMed] [Google Scholar]
- 24.Goehring, N. W., M. D. Gonzalez, and J. Beckwith. 2006. Premature targeting of cell division proteins to midcell reveals hierarchies of protein interactions involved in divisome assembly. Mol. Microbiol. 6133-45. [DOI] [PubMed] [Google Scholar]
- 25.Goehring, N. W., F. Gueiros-Filho, and J. Beckwith. 2005. Premature targeting of a cell division protein to midcell allows dissection of divisome assembly in Escherichia coli. Genes Dev. 19127-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grantcharova, N., U. Lustig, and K. Flardh. 2005. Dynamics of FtsZ assembly during sporulation in Streptomyces coelicolor A3(2). J. Bacteriol. 1873227-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grantcharova, N., W. Ubhayasekera, S. L. Mowbray, J. R. McCormick, and K. Flardh. 2003. A missense mutation in ftsZ differentially affects vegetative and developmentally controlled cell division in Streptomyces coelicolor A3(2). Mol. Microbiol. 47645-656. [DOI] [PubMed] [Google Scholar]
- 28.Hara, H., S. Yasuda, K. Horiuchi, and J. T. Park. 1997. A promoter for the first nine genes of the Escherichia coli mra cluster of cell division and cell envelope biosynthesis genes, including ftsI and ftsW. J. Bacteriol. 1795802-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikeda, M., T. Sato, M. Wachi, H. K. Jung, F. Ishino, Y. Kobayashi, and M. Matsuhashi. 1989. Structural similarity among Escherichia coli FtsW and RodA proteins and Bacillus subtilis SpoVE protein, which function in cell division, cell elongation, and spore formation, respectively. J. Bacteriol. 1716375-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang, H., and K. E. Kendrick. 2000. Cloning and characterization of the gene encoding penicillin-binding protein A of Streptomyces griseus. FEMS Microbiol. Lett. 19363-68. [DOI] [PubMed] [Google Scholar]
- 31.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 32.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305567-580. [DOI] [PubMed] [Google Scholar]
- 33.Kwak, J., A. J. Dharmatilake, H. Jiang, and K. E. Kendrick. 2001. Differential regulation of ftsZ transcription during septation of Streptomyces griseus. J. Bacteriol. 1835092-5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lara, B., and J. A. Ayala. 2002. Topological characterization of the essential Escherichia coli cell division protein FtsW. FEMS Microbiol. Lett. 21623-32. [DOI] [PubMed] [Google Scholar]
- 35.Letek, M., E. Ordonez, M. Fiuza, P. Honrubia-Marcos, J. Vaquera, J. A. Gil, D. Castro, and L. M. Mateos. 2007. Characterization of the promoter region of ftsZ from Corynebacterium glutamicum and controlled overexpression of FtsZ. Int. Microbiol. 10271-282. [PubMed] [Google Scholar]
- 36.Marston, A. L., H. B. Thomaides, D. H. Edwards, M. E. Sharpe, and J. Errington. 1998. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes. Dev. 123419-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCormick, J. R., and R. Losick. 1996. Cell division gene ftsQ is required for efficient sporulation but not growth and viability in Streptomyces coelicolor A3(2). J. Bacteriol. 1785295-5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCormick, J. R., E. P. Su, A. Driks, and R. Losick. 1994. Growth and viability of Streptomyces coelicolor mutant for the cell division gene ftsZ. Mol. Microbiol. 14243-254. [DOI] [PubMed] [Google Scholar]
- 39.Mercer, K. L., and D. S. Weiss. 2002. The Escherichia coli cell division protein FtsW is required to recruit its cognate transpeptidase, FtsI (PBP3), to the division site. J. Bacteriol. 184904-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molle, V., W. J. Palframan, K. C. Findlay, and M. J. Buttner. 2000. WhiD and WhiB, homologous proteins required for different stages of sporulation in Streptomyces coelicolor A3(2). J. Bacteriol. 1821286-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen, L., N. Scherr, J. Gatfield, A. Walburger, J. Pieters, and C. J. Thompson. 2007. Antigen 84, an effector of pleiomorphism in Mycobacterium smegmatis. J. Bacteriol. 1897896-7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noens, E. E., V. Mersinias, J. Willemse, B. A. Traag, E. Laing, K. F. Chater, C. P. Smith, H. K. Koerten, and G. P. van Wezel. 2007. Loss of the controlled localization of growth stage-specific cell-wall synthesis pleiotropically affects developmental gene expression in an ssgA mutant of Streptomyces coelicolor. Mol. Microbiol. 641244-1259. [DOI] [PubMed] [Google Scholar]
- 43.Rajagopalan, M., E. Maloney, J. Dziadek, M. Poplawska, H. Lofton, A. Chauhan, and M. V. Madiraju. 2005. Genetic evidence that mycobacterial FtsZ and FtsW proteins interact, and colocalize to the division site in Mycobacterium smegmatis. FEMS Microbiol. Lett. 2509-17. [DOI] [PubMed] [Google Scholar]
- 44.Ramos, A., M. P. Honrubia, N. Valbuena, J. Vaquera, L. M. Mateos, and J. A. Gil. 2003. Involvement of DivIVA in the morphology of the rod-shaped actinomycete Brevibacterium lactofermentum. Microbiology 1493531-3542. [DOI] [PubMed] [Google Scholar]
- 45.Real, G., A. Fay, A. Eldar, S. M. Pinto, A. O. Henriques, and J. Dworkin. 2008. Determinants for the subcellular localization and function of a nonessential SEDS protein. J. Bacteriol. 190363-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Redenbach, M., H. M. Kieser, D. Denapaite, A. Eichner, J. Cullum, H. Kinashi, and D. A. Hopwood. 1996. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol. Microbiol. 2177-96. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 48.Scheffers, D. J., C. Robichon, G. J. Haan, T. den Blaauwen, G. Koningstein, E. van Bloois, J. Beckwith, and J. Luirink. 2007. Contribution of the FtsQ transmembrane segment to localization to the cell division site. J. Bacteriol. 1897273-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwedock, J., J. R. McCormick, E. R. Angert, J. R. Nodwell, and R. Losick. 1997. Assembly of the cell division protein FtsZ into ladder-like structures in the aerial hyphae of Streptomyces coelicolor. Mol. Microbiol. 25847-858. [DOI] [PubMed] [Google Scholar]
- 50.Thanky, N. R., D. B. Young, and B. D. Robertson. 2007. Unusual features of the cell cycle in mycobacteria: polar-restricted growth and the snapping-model of cell division. Tuberculosis 87231-236. [DOI] [PubMed] [Google Scholar]
- 51.Wehmeier, U. F. 1995. New multifunctional Escherichia coli-Streptomyces shuttle vectors allowing blue-white screening on XGal plates. Gene 165149-150. [DOI] [PubMed] [Google Scholar]
- 52.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33103-119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.