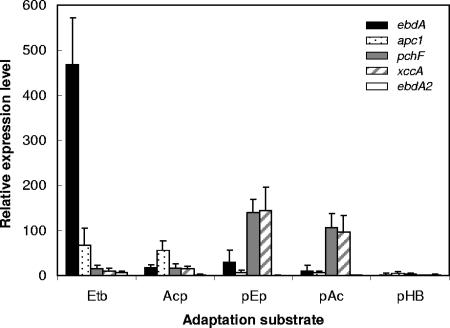
FIG. 5.
Relative expression levels of ebdA, apc1, pchF, and xccA in substrate-adapted cells of “A. aromaticum” strain EbN1. The selected genes encode catalytic subunits ethylbenzene dehydrogenase (ebdA), acetophenone carboxylase (apc1), predicted p-ethylphenol methylhydroxylase (pchF), predicted p-hydroxyacetophenone carboxylase (xccA), and paralogous catalytic subunit of ethylbenzene dehydrogenase (ebdA2). Benzoate-adapted cells were used as a reference state, and bcrC (encoding the γ-subunit of benzoyl-CoA reductase) served as a reference gene. Relative expression levels were determined by real-time RT-PCR. Abbreviations of adaptation substrates: Etb, ethylbenzene; Acp, acetophenone; pEp, p-ethylphenol; pAc, p-acetophenone; pHB, p-hydroxybenzoate.