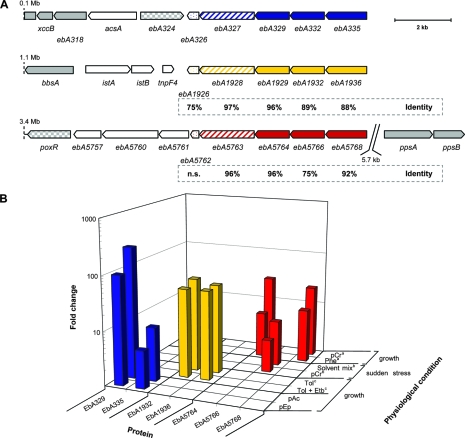
FIG. 6.
Gene products possibly involved in solvent stress tolerance of “A. aromaticum” strain EbN1. (A) Chromosomal gene locations are indicated by their nucleotide positions. Genes encoding stress-related proteins are highlighted in blue, yellow, and red. Hypothetical proteins, filled; predicted efflux pump, hatched; and predicted universal stress protein, dotted. Amino acid sequence identities of gene products (referring to EbA326-335) are indicated below the paralogous genes (n.s., no significant similarity). Genes encoding proteins of aromatic compound catabolism are highlighted in gray (filled), and respective regulatory proteins are checkered. (B) Changes (n-fold) in abundances of identified hypothetical proteins (EbA329-335, EbA1932-1936, and EbA5764-5768) during anaerobic growth shock with different aromatic substrates, as determined by 2D DIGE. Substrate abbreviations: pEp, p-ethylphenol; pAc, p-acetophenone; Tol, toluene; Etb, ethylbenzene; pCr, p-cresol; Phe, phenol. “a,” anaerobic growth with benzoate represents the reference state. No upregulation during anaerobic growth with phenylalanine, phenylacetate, benzyl alcohol, benzaldehyde, p-hydroxybenzoate, m-hydroxybenzoate, and o-aminobenzoate (52). “b,” anaerobic growth with succinate represents the reference state (49). “c,” anaerobic growth with benzoate represents the reference state. No induction during anaerobic growth with ethylbenzene (26).