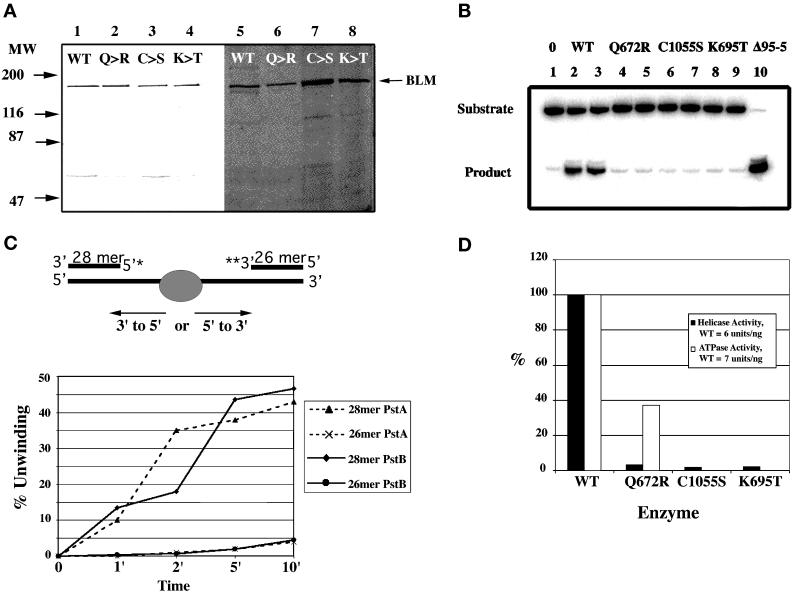
Figure 4.
Purification and DNA helicase activity of BLM. (A) Isolation of normal and missense proteins of BLM. The left panel (lanes 1–4) contains isolated BLM proteins transferred to a PVDF membrane and reacted with the BLM antisera. Equal amounts of recovered protein are loaded in each lane. The right panel is a silver-stained polyacrylamide gel of the isolated proteins (lanes 5–8). Equal volumes of equivalent fractions from each preparation are loaded to show relative recoveries of the different proteins from yeast strain AMR 61. Molecular weight markers are indicated on the left. Lanes 1 and 5 contain normal BLM; lanes 2 and 6 contain Q672R missense protein; lanes 3 and 7 contain C1055S missense protein; lanes 4 and 8 contain K695T missense protein. (B) Helicase activity of the normal and missense BLM proteins. Displacement activity is measured using a 3′ end-labeled oligonucleotide 54 bases long annealed to ssMp18 DNA. Lanes marked 0 and Δ95–5 are the substrate (no enzyme) and the product (substrate heated at 95°C for 5 min). Each reaction contains 1 ng of recovered protein: lanes 2 and 3, normal BLM; lanes 4 and 5, missense Q672R BLM; lanes 6 and 7 missense C1055S BLM; lanes 8 and 9 missense K695T BLM. (C) Polarity of the BLM helicase. The direction of movement of the BLM helicase was evaluated using a substrate digested with PstI that creates two oligonucleotides of different sizes at opposite ends of the long ssMp18 DNA. In experiment PstA, both ends of the same substrate molecule were labeled (5′ and 3′) before digestion with PstI, and in PstB two different substrate molecules were labeled either at the 5′ or the 3′ end, digested with PstI and equal amounts of each mixed together in the reaction tubes. Assays were quantitated with a Molecular Dynamics PhosphorImager. (D) Comparison of helicase activities of normal and missense BLM proteins. Displacement activity and DNA-dependent ATPase specific activity is shown as a percentage of normal protein activity.