Abstract
Dermatophytes are fungi that belong to three genera: Epidermophyton, Microsporum, and Trichophyton. Identification of dermatophyte species is essential for appropriate diagnosis and treatment of dermatophytosis. Routine identification depends on macroscopic and microscopic morphology, which is time-consuming and does not identify dermatophyte strains. In this study, two PCR-based methods were compared for their abilities to identify 21 dermatophyte isolates obtained from Egyptian patients to the species and strain levels. The first method employed a two-step method: PCR amplification, using ITS1 and ITS4 as primers, followed by restriction enzyme digestion using the endonuclease MvaI. The second method employed a one-step approach employing the repetitive oligonucleotide (GACA)4 as a primer. Dermatophyte strains were also identified using a conventional culture method. Our results showed that the conventional culture method identified four species: Microsporum canis, Trichophyton mentagrophytes, Trichophyton rubrum, and Trichophyton violaceum. Moreover, both PCR methods agreed with the diagnosis made using the conventional approach. Furthermore, ITS1/ITS4-based PCR provided no strain differentiation, while (GACA)4-based PCR identified different varieties among the T. mentagrophytes isolates. Taken together, our results suggest that (GACA)4-based PCR has utility as a simple and rapid method for identification of dermatophyte species as well as utility for differentiation of T. mentagrophytes variants.
Dermatophytes comprise a group of related fungi that belong to three genera, Epidermophyton, Microsporum, and Trichophyton, each of which includes several recognized species. These fungi are keratinophylic, as they infect the superficial keratinized tissues (skin, hair, and nails) of humans and animals (16), and can cause cutaneous mycoses which are called dermatophytoses, tinea, or ringworm infections. Health care costs associated with management of these mycoses are high (2, 14). Moreover, dermatophytoses are widespread and increasing in prevalence on a global scale and the recent increase in their incidence has been attributed to the increase of immunocompromised states, such as those associated with AIDS, diabetes mellitus, organ transplantation, and the use of corticosteroids and antineoplastic agents (3, 5, 11, 17).
The identification of dermatophyte species is essential for appropriate diagnosis and treatment of dermatophytosis. As the dermatophytes were reported to cause outbreaks of infection, especially in closed communities (15), their identification to the species as well as strain levels has a great epidemiological value in the investigations of such outbreaks with regard to identifying the sources of infections and establishing plans to manage and control them.
Routine laboratory procedures for the identification of dermatophytes rely on culturing of these fungi on appropriate growth media, followed by examination of the gross morphological characters of their colonies (e.g., rate of growth, colony topography, and pigmentation of the surface and reverse sides) as well as microscopic morphology (e.g., shape and size of macroconidia, microconidia, and hyphae). Further identification characteristics include nutritional requirements (such as vitamin and amino acid utilization), temperature tolerance, urease production, in vitro hair perforation, etc. (16). Although culture-based identification is specific and sensitive, it is time-consuming since some species need up to 2 to 3 weeks before diagnostic characteristics are fully developed in culture media. Additionally, many dermatophyte strains often develop atypical characteristics.
Many molecular approaches have been applied for identification of different dermatophyte species and strains. Such approaches are considered more stable and precise than those using phenotypic characteristics (7). One such approach employs PCR technology, which is simple, rapid, and able to generate species-specific DNA polymorphisms with many dermatophyte species on the basis of characteristic band patterns detected by agarose gel electrophoresis (4, 8).
Jackson et al. (8) used the internal transcribed spacer (ITS) region of ribosomal DNA as a target for PCR amplification using the ITS1 and ITS4 primers, followed by MvaI restriction enzyme digestion, for identification of 17 dermatophyte species. This method produced unique fragment patterns for most dermatophytic species studied but could not distinguish between closely related species, such as Trichophyton rubrum and Trichophyton soudanense or Trichophyton quinckeanum and Trichophyton schoenleinii. Faggi et al. (4) used a one-step PCR-based approach employing the simple repetitive oligonucleotide (GACA)4 as a single primer for identification of species of dermatophytes. Their data showed that this simple primer was able to amplify all the studied dermatophytes with production of species-specific PCR profiles. However, no head-to-head study comparing these two PCR-based approaches has been undertaken.
The present study aimed at comparing these two molecular PCR-based methods for identification of 21 dermatophyte strains isolated from patients in Egypt. The purpose of this study was to identify which of these methods is easier to perform and can differentiate between dermatophyte species as well as strains.
MATERIALS AND METHODS
Dermatophyte isolates.
Twenty-one clinical dermatophyte strains were recovered from Egyptian patients with different clinical types of dermatophytosis. Skin, hair, and nail samples were collected from the patients and cultured on Mycosel agar (BBL; Becton, Dickinson & Co., MD). These clinical isolates were identified to the species level by using routine phenotypic methods, including colony morphology, microscopy, physiologic, and biochemical tests. All of the isolated strains were maintained on Mycosel agar slants. Four reference strains, Trichophyton rubrum ATCC 28188, Trichophyton mentagrophytes ATCC 9533, Trichophyton violaceum MRL 2135, and Microsporum canis MRL 2117, were obtained from the culture collection at the Center for Medical Mycology, Cleveland, OH.
DNA extraction.
Fungal isolates were grown in 50 ml Sabouraud dextrose broth (BBL; Becton, Dickinson & Co., MD) and incubated with shaking at 30°C for 7 days for all strains except the T. violaceum strains, which required incubation for 2 weeks. Fungal growth was harvested by filtration using a 0.22-μm Stericup filter (Millipore Corporation, Billerica, MA) and washed several times by 0.1 M MgCl2. The specimens were transferred to sterile, prechilled mortars, then liquid nitrogen was added, and the specimens were ground finely with a pestle. The resulting powder was aliquoted into sterile, 1.5-ml microcentrifuge tubes. The powder specimens that could not be processed immediately were frozen at −20°C. Fungal DNA was extracted using a MasterPure yeast DNA purification kit (Epicentre Biotechnologies, Madison, WI) according to the manufacturer's instructions. The extracted DNA was treated with RNase A, and then DNA was extracted with phenol-chloroform (MP Biomedicals, Inc., Solon, OH) and finally with chloroform (Fisher scientific, Fair lawn, NJ), precipitated with ice-cold absolute ethanol, washed with 1 ml of 70% ethanol, air dried, and resuspended in 50 μl of Tris-EDTA buffer.
PCR method using the ITS1 and ITS4 primers.
Amplification reactions were carried out with volumes of 100 μl containing reaction buffer (50 mM KCl, 10 mM Tris-HCl [pH 9.0], 0.1% Triton X-100, 1.5 mM MgCl2, deoxynucleoside triphosphate mix [0.2 mM each of dATP, dCTP, dGTP, and dTTP], 30 pmol each of primers ITS1 [5′ TCCGTAGGTGAACCTGCGG 3′] and ITS4 [5′ TCCTCCGCTTATTGATATGC 3′] [Operon Biotechnologies, Inc., Huntsville, AL], 5 U of Taq polymerase [Roche Diagnostics GmbH, Mannheim, Germany], and approximately 10 ng of template DNA, made up to a total volume of 100 μl with pure, sterile double-distilled water). The PCR cycling conditions were 35 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 2 min, followed by an extension step of 72°C for 10 min. PCR was carried out using a thermal cycler (iCycler; Bio-Rad, Hercules, CA). PCR products were purified using a QIAquick PCR purification kit (Qiagen Sciences, MD) according to the manufacturer's directions. The purified PCR products were digested with the restriction endonuclease enzyme MvaI (Takara Bio., Inc., Otsu, Shiga, Japan), which recognizes the sequence 5′ CC (T/A) GG 3′. The resulting products were separated in 2% agarose gels and 1× Tris-acetate-EDTA buffer and stained with ethidium bromide, and then images were captured using the Versadoc imaging system (Bio-Rad, CA).
PCR method using the (GACA)4 primer.
Amplification reactions were carried out with volumes of 50 μl containing reaction buffer [50 mM KCl, 10 mM Tris-HCl (pH 9.0), 2.5 mM MgCl2, deoxynucleoside triphosphate mix (0.2 mM each of dATP, dCTP, dGTP, and dTTP), 160 ng of the (GACA)4 primer (Operon Biotechnologies), 2.5 U of Taq polymerase (Roche Diagnostics), and approximately 25 ng of template DNA, made up to a total volume 50 μl with pure, sterile double-distilled water]. PCR was carried out for 39 cycles of denaturation at 93°C for 1 min, annealing at 50°C for 1 min, and extension at 72°C for 1 min, followed by a final extension step at 72°C for 7 min. The resulting PCR products were separated in 1% agarose gels in 0.5× Tris-borate-EDTA buffer and stained with ethidium bromide, and then images were obtained as described above.
RESULTS
Culture results.
The 21 isolated dermatophyte strains were found to belong to four species: T. violaceum (8 strains), T. rubrum (4 strains), T. mentagrophytes (5 strains, comprising 2 T. mentagrophytes var. mentagrophytes and 3 T. mentagrophytes var. interdigitale strains), and M. canis (4 strains). All strains of T. violaceum and M. canis were isolated from patients with tinea capitis or tinea corporis, while T. rubrum and T. mentagrophytes strains were obtained from patients with tinea corporis, tinea cruris, or tinea unguium. The clinical samples used to isolate these dermatophytes varied with the type of infection: hair for tinea capitis, skin scrapings for tinea corporis and tinea cruris, and nail scrapings for tinea unguium (Table 1).
TABLE 1.
Demographic information for the isolated dermatophyte strains
| Isolate no. | Type of dermatophyte infection | Clinical specimen | Patient
|
Dermatophyte isolated | |
|---|---|---|---|---|---|
| Sex | Age (yrs) | ||||
| 1 | Tinea capitis | Hair | Male | 7 | M. canis |
| 2 | Tinea cruris | Skin scrapings | Female | 40 | T. rubrum |
| 3 | Tinea corporis | Skin scrapings | Female | 48 | T. rubrum |
| 4 | Tinea capitis | Hair | Male | 6.5 | T. violaceum |
| 5 | Tinea capitis | Hair | Male | 9 | T. violaceum |
| 6 | Tinea capitis | Hair | Male | 4 | T. violaceum |
| 7 | Tinea capitis | Hair | Male | 4 | T. violaceum |
| 8 | Tinea capitis | Hair | Male | 6 | T. violaceum |
| 9 | Tinea corporis | Skin scrapings | Male | 6 | T. violaceum |
| 10 | Tinea capitis | Hair | Female | 12 | M. canis |
| 11 | Tinea capitis | Hair | Male | 10 | M. canis |
| 12 | Tinea corporis | Skin scrapings | Male | 31 | T. mentagrophytes var. mentagrophytes |
| 13 | Tinea cruris | Skin scrapings | Male | 39 | T. rubrum |
| 14 | Tinea unguium | Nail scrapings | Female | 48 | T. rubrum |
| 15 | Tinea corporis | Skin scrapings | Male | 4 | M. canis |
| 16 | Tinea corporis | Skin scrapings | Male | 35 | T. violaceum |
| 17 | Tinea cruris | Skin scrapings | Male | 17.5 | T. mentagrophytes var. interdigitale |
| 18 | Tinea capitis | Hair | Female | 6 | T. violaceum |
| 19 | Tinea cruris | Skin scrapings | Male | 20 | T. mentagrophytes var. mentagrophytes |
| 20 | Tinea corporis | Skin scrapings | Male | 28 | T. mentagrophytes var. interdigitale |
| 21 | Tinea unguium | Nail scrapings | Female | 43 | T. mentagrophytes var. interdigitale |
Dermatophyte identification using ITS-based PCR.
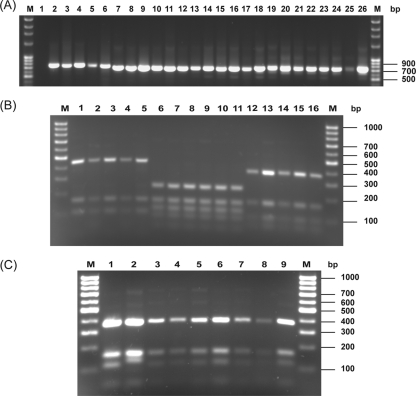
Analysis of the ITS regions of ribosomal DNA is a simple and reproducible molecular tool for identification of dermatophyte species. To determine the agreement of PCR-based methods with culture-based techniques, we performed PCR-based identification using PCR with the ITS1/ITS4 primer set, followed by MvaI digestion. This primer set amplified the ITS I, 5.8S, and ITS II regions of the ribosomal DNA in all 21 strains tested, resulting in amplified products of approximately 690 bp in the T. violaceum, T. rubrum, and T. mentagrophytes species. For the M. canis species, the size of the amplified product was approximately 740 bp (Fig. 1A). MvaI digestion of these amplified products from each of the four isolated species revealed unique restriction patterns, with no intraspecies variation (Fig. 1B and C). M. canis isolates showed three band patterns, ranging from 100 bp to 500 bp in size, with a marked size difference between the largest and middle bands. On the other hand, T. mentagrophytes strains had a more complex pattern that contained five bands within a narrow size range (50 bp to 280 bp), with no variation between the two tested T. mentagrophytes varieties. Both T. rubrum and T. violaceum isolates resulted in four bands, with sizes ranging between 50 bp and 400 bp, but there was a difference noted between these two species in the distribution of those bands, as the middle two bands were closer in size in T. violaceum than in T. rubrum strains.
FIG. 1.
Agarose gel electrophoresis. (A) PCR products of the ITS I, 5.8S, and ITS II regions of the four phenotypically identified species. Lanes: M, molecular weight marker (Fisher Scientific International, Inc.); 1, negative control (no template DNA); 2, M. canis MRL 2117; 3 to 6, M. canis clinical strains; 7, T. mentagrophytes ATCC 9533; 8 to 12, T. mentagrophytes clinical strains; 13, T. rubrum ATCC 28188; 14 to 17, T. rubrum clinical strains; 18, T. violaceum MRL 2135; 19 to 26, T. violaceum clinical strains. (B) MvaI restriction products of M. canis, T. mentagrophytes, and T. rubrum species. Lanes: M, molecular weight marker (Fisher Scientific International, Inc.); 1, M. canis MLR 2117; 2 to 5 M. canis clinical strains; 6, T. mentagrophytes ATCC 9533; 7 to 11, T. mentagrophytes clinical strains; 12, T. rubrum ATCC 28188; 13 to 16, T. rubrum clinical strains. (C) MvaI restriction products of T. violaceum isolates. Lanes: M, molecular weight marker (Fisher Scientific International, Inc.); 1, T. violaceum MRL 2135; 2 to 9, T. violaceum clinical strains.
Identification of dermatophytes by use of (GACA)4 primer-based PCR.
The second PCR-based method used in this study utilized the short oligonucleotide (GACA)4 as a primer for identification of the tested dermatophyte isolates. All of the studied strains were amplified with this simple, repetitive primer, and the numbers of the resulting PCR bands ranged from 4 to 11 (size range, 300 bp to 2,500 bp). All T. violaceum strains gave nearly the same pattern, which consisted of three bright bands (approximately 600 bp, 900 bp, and 1,000 bp) and one to three additional faint bands with sizes ranging from 1,600 bp to 2,500 bp (Fig. 2A). T. rubrum strains could be distinguished from T. violaceum strains by the sizes of the three largest bands (1,900 to 2,500 bp) (Fig. 2B).
FIG. 2.
Agarose gel electrophoresis for PCR using the (GACA)4 primer. (A) T. violaceum strains. Lanes: M, molecular weight marker (Fisher Scientific International, Inc.); 1, negative control (no template DNA); 2, T. violaceum MRL 2135; 3 to 10, T. violaceum clinical strains. (B) T. rubrum strains. Lanes: M, molecular weight marker (Fisher Scientific International, Inc.); 1, negative control (no template DNA); 2, T. rubrum ATCC 28188; 3 to 6, T. rubrum clinical strains. (C) T. mentagrophytes strains. Lanes: M, molecular weight marker (Fisher Scientific International, Inc.); 1, negative control (no template DNA); 2, T. mentagrophytes ATCC 9533; 3 to 7, T. mentagrophytes clinical strains. (D) M. canis strains. Lanes: M, molecular weight marker (Fisher Scientific International, Inc.); 1, negative control (no template DNA); 2, M. canis MRL 2117; 3 to 6, M. canis clinical strains.
For the five T. mentagrophytes strains (two T. mentagrophytes var. mentagrophytes and three T. mentagrophytes var. interdigitale strains), three profiles were observed (Fig. 2C). The first profile (Fig. 2C, lane 3) consisted of six bands, ranging from 300 bp to 1,300 bp in size, with one strong band of 600 bp, and five faint bands (with one large fragment of 1,300 bp). The second and third profiles (Fig. 2C, lane 4 and lanes 5 to 7, respectively) were more complex, comprising eight or nine bands, between 300 bp and approximately 1,800 bp in size. The last two profiles differed by two bands of approximately 1,300 and 1,400 bp. Both the first and the second profiles were represented in T. mentagrophytes var. mentagrophytes strains (one strain to each profile), while the third profile was represented in the three T. mentagrophytes var. interdigitale strains. Moreover, (GACA)4-based PCR of M. canis strains revealed the most complex profiles, with up to 11 bands, ranging from 600 bp to 2,500 bp in size. There was no intraspecies variation among M. canis isolates, all of which had the same band pattern (Fig. 2D).
DISCUSSION
Molecular techniques are more beneficial for dermatophyte identification as they are rapid and more sensitive. Moreover, these methods rely on genetic makeup, which is more constant than phenotypic characterization, and they can identify atypical dermatophytes that could not be identified by culture-based techniques (6). These genotypic approaches can identify the dermatophytes to the species as well as the strain levels (1, 8, 9, 18).
In this study, we compared the utilities of two molecular PCR-based methods (with two different primer sets) for identification of four dermatophyte species (M. canis, T. mentagrophytes, T. rubrum, and T. violaceum) isolated from patients with dermatophytosis.
The first method employed PCR to amplify ITS regions by using the ITS1/ITS4 primer set, followed by restriction fragment length polymorphism (RFLP) analysis of the amplified products by use of the MvaI restriction enzyme. PCR amplification resulted in products of 690 bp for T. mentagrophytes, T. rubrum, and T. violaceum isolates and 740 bp for M. canis. Subsequent MvaI digestion of these products identified the studied species in complete agreement with the culture-based identification, and the obtained profiles were unique to each of the studied species, but no strain variation was detected in any of the studied species by using this method. Our results are in agreement with those reported by Jackson et al. (8), who found that PCR-RFLP of the ITS region was a useful molecular tool for the identification of dermatophytes to the species level. All of these findings indicate that the dermatophyte ITS regions are conserved and can be used as a useful marker for dermatophyte species differentiation.
On the other hand, the absence of intraspecies variation revealed by this method suggested that strain-specific variations of the studied species are not located in the ITS regions, but instead, these variations may reside in other regions of the DNA, such as the nontranscribed spacer (NTS) region. Jackson et al. (9) studied this NTS region for strain identification of T. rubrum, and they found that the NTS region had two tandemly repetitive subelements that provided strain-specific patterns for T. rubrum. In this regard, the sequencing of the NTS region of T. mentagrophytes var. interdigitale revealed three polymorphic subrepeat loci and PCR fingerprinting of these loci for 42 random isolates of this species revealed 19 individual strain profiles (10).
In the second PCR-based method, we used the short oligonucleotide (GACA)4 as a single primer. This primer has been shown to be a useful tool in molecular identification of dermatophytes (4, 13). In the current study, we successfully identified the tested isolates to the species level in full agreement with both the culture-based and the ITS-based PCR-RFLP methods. Our study also revealed that the (GACA)4-based PCR band profiles were more complex than the PCR-RFLP profiles.
The obtained profiles were characteristic of each species tested in this study, but marked similarity was observed between the profiles of the T. rubrum and T. violaceum strains, which can be explained by the close relatedness of these two species. This close relationship was previously demonstrated by Ohst et al. (12), who analyzed the population structures of both species (T. rubrum and T. violaceum) by using a microsatellite marker, T1. This marker, which was developed by an enrichment technique for microsatellites and contained the (GT)8-10 repeat, was found to specifically amplify both species.
In our study, the profile of M. canis was more complex, as it contained a large number of bands and a wide range of band sizes among different isolates, but all patterns were similar, with no strain pattern variation. In spite of the different geographical origins and sources of the M. canis isolates used in our study and those used in the Faggi et al. study (4), the resultant profiles from both studies were in concordance, with up to 11 bands, ranging in size from approximately 600 bp to 2,500 bp. This agreement supports the contention that (GACA)4-based PCR is a reproducible method.
The T. mentagrophytes isolates exhibited three distinct band profiles, with a range of six to nine bands of various sizes (approximately 300 bp to 1,800 bp). These profiles were markedly different from those of the other species tested in this study. Also, (GACA)4-based PCR revealed an association between the tested T. mentagrophytes varieties and the obtained band profiles, as shown by the presence of three different profiles among two varieties. T. mentagrophytes var. mentagrophytes strains showed two different profiles, while all three strains of T. mentagrophytes var. interdigitale showed a distinct, third profile. The band profiles obtained in our study matched those reported by Faggi et al. (4). In this regard, we found that both the first and the second profiles in our study matched the profiles for T. mentagrophytes isolates of unspecified variety described by Faggi et al. (4), while the third profile found in our study matched the pattern of T. mentagrophytes var. interdigitale described by these investigators. Therefore, the (GACA)4-based method has utility for the identification of species as well as varieties of T. mentagrophytes. Moreover, the difference between the profiles of the T. mentagrophytes and T. rubrum species can provide a more helpful tool that can be used in differentiation of these two species, which may otherwise be easily confused when they are identified by routine culture-based methods, due to marked similarities in their phenotypic features.
Taken together, our results showed that (GACA)4-based PCR is a simple, easy, rapid, and reproducible molecular technique that has utility for identification of dermatophyte fungi to the species level. Moreover, this method also has potential value in identification of T. mentagrophytes variants. Evaluation of this method by use of a larger panel of dermatophyte species and strains is warranted.
Acknowledgments
This study was performed as part of the M.D. thesis of A.S., with partial support from the Ministry of Higher Education, Egypt. Funds were also provided by the Center for Medical Mycology, Case Western Reserve University, Cleveland, OH.
Footnotes
Published ahead of print on 25 June 2008.
REFERENCES
- 1.Abdel-Rahman, S. M., S. Simon, K. J. Wright, L. Ndjountche, and A. Gaedigk. 2006. Tracking Trichophyton tonsurans through a large urban child care center: defining infection prevalence and transmission patterns by molecular strain typing. Pediatrics 1182365-2373. [DOI] [PubMed] [Google Scholar]
- 2.Drake, L. A., S. M. Dinehart, E. R. Farmer, R. W. Goltz, G. F. Graham, M. K. Hardinsky, C. W. Lewis, D. M. Pariser, J. W. Skouge, S. B. Webster, D. C. Whitaker, B. Butler, B. J. Lowery, B. E. Elewski, M. L. Elgart, P. H. Jacobs, J. L. Lesher, Jr., R. K. Scher, et al. 1996. Guidelines of care for superficial mycotic infections of the skin: tinea corporis, tinea cruris, tinea faciei, tinea manuum, and tinea pedis. J. Am. Acad. Dermatol. 34282-286. [DOI] [PubMed] [Google Scholar]
- 3.Faergemann, J., and R. Baran. 2003. Epidemiology, clinical presentation and diagnosis of onychomycosis. Br. J. Dermatol. 149(Suppl 65)1-4. [DOI] [PubMed] [Google Scholar]
- 4.Faggi, E., G. Pini, E. Campisi, C. Bertellini, E. Difonzo, and F. Mancianti. 2001. Application of PCR to distinguish common species of dermatophytes. J. Clin. Microbiol. 393382-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghannoum, M. A., R. A. Hajjeh, R. Scher, N. Konnikov, A. K. Gupta, R. Summerbell, S. Sullivan, R. Daniel, P. Krusinski, P. Fleckman, P. Rich, R. Odom, R. Aly, D. Pariser, M. Zaiac, G. Rebell, J. Lesher, B. Gerlach, G. F. Ponce-De-Leon, A. Ghannoum, J. Warner, N. Isham, and B. Elewski. 2000. A large-scale North American study of fungal isolates from nails: the frequency of onychomycosis, fungal distribution, and antifungal susceptibility patterns. J. Am. Acad. Dermatol. 43641-648. [DOI] [PubMed] [Google Scholar]
- 6.Graser, Y., M. el Fari, W. Presber, W. Sterry, and H. J. Tietz. 1998. Identification of common dermatophytes (Trichophyton, Microsporum, Epidermophyton) using polymerase chain reactions. Br. J. Dermatol. 138576-582. [DOI] [PubMed] [Google Scholar]
- 7.Gräser, Y., A. F. Kuijpers, W. Presber, and G. S. de Hoog. 2000. Molecular taxonomy of the Trichophyton rubrum complex. J. Clin. Microbiol. 383329-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson, C. J., R. C. Barton, and E. G. Evans. 1999. Species identification and strain differentiation of dermatophyte fungi by analysis of ribosomal-DNA intergenic spacer regions. J. Clin. Microbiol. 37931-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson, C. J., R. C. Barton, S. L. Kelly, and E. G. Evans. 2000. Strain identification of Trichophyton rubrum by specific amplification of subrepeat elements in the ribosomal DNA nontranscribed spacer. J. Clin. Microbiol. 384527-4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson, C. J., T. Mochizuki, and R. C. Barton. 2006. PCR fingerprinting of Trichophyton mentagrophytes var. interdigitale using polymorphic subrepeat loci in the rDNA nontranscribed spacer. J. Med. Microbiol. 551349-1355. [DOI] [PubMed] [Google Scholar]
- 11.Mirmirani, P., N. A. Hessol, T. A. Maurer, T. G. Berger, P. Nguyen, A. Khalsa, A. Gurtman, S. Micci, M. Young, S. Holman, S. J. Gange, and R. M. Greenblatt. 2001. Prevalence and predictors of skin disease in the Women's Interagency HIV Study (WIHS). J. Am. Acad. Dermatol. 44785-788. [DOI] [PubMed] [Google Scholar]
- 12.Ohst, T., S. de Hoog, W. Presber, V. Stavrakieva, and Y. Graser. 2004. Origins of microsatellite diversity in the Trichophyton rubrum-T. violaceum clade (dermatophytes). J. Clin. Microbiol. 424444-4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roque, H. D., R. Vieira, S. Rato, and M. Luz-Martins. 2006. Specific primers for rapid detection of Microsporum audouinii by PCR in clinical samples. J. Clin. Microbiol. 444336-4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith, E. S., A. B. Fleischer, Jr., and S. R. Feldman. 1998. Nondermatologists are more likely than dermatologists to prescribe antifungal/corticosteroid products: an analysis of office visits for cutaneous fungal infections, 1990-1994. J. Am. Acad. Dermatol. 3943-47. [DOI] [PubMed] [Google Scholar]
- 15.Stiller, M. J., W. P. Klein, R. I. Dorman, and S. Rosenthal. 1992. Tinea corporis gladiatorum: an epidemic of Trichophyton tonsurans in student wrestlers. J. Am. Acad. Dermatol. 27632-633. [DOI] [PubMed] [Google Scholar]
- 16.Weitzman, I., and R. C. Summerbell. 1995. The dermatophytes. Clin. Microbiol. Rev. 8240-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodfolk, J. A. 2005. Allergy and dermatophytes. Clin. Microbiol. Rev. 1830-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu, J., Z. Wan, W. Chen, W. Wang, and R. Li. 2004. Molecular typing study of the Microsporum canis strains isolated from an outbreak of tinea capitis in a school. Mycopathologia 15737-41. [DOI] [PubMed] [Google Scholar]